Gene ontology analysis of pairwise genetic associations in two genome-wide studies of sporadic ALS
- PMID: 22839596
- PMCID: PMC3463436
- DOI: 10.1186/1756-0381-5-9
Gene ontology analysis of pairwise genetic associations in two genome-wide studies of sporadic ALS
Abstract
Background: It is increasingly clear that common human diseases have a complex genetic architecture characterized by both additive and nonadditive genetic effects. The goal of the present study was to determine whether patterns of both additive and nonadditive genetic associations aggregate in specific functional groups as defined by the Gene Ontology (GO).
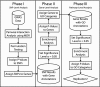
Results: We first estimated all pairwise additive and nonadditive genetic effects using the multifactor dimensionality reduction (MDR) method that makes few assumptions about the underlying genetic model. Statistical significance was evaluated using permutation testing in two genome-wide association studies of ALS. The detection data consisted of 276 subjects with ALS and 271 healthy controls while the replication data consisted of 221 subjects with ALS and 211 healthy controls. Both studies included genotypes from approximately 550,000 single-nucleotide polymorphisms (SNPs). Each SNP was mapped to a gene if it was within 500 kb of the start or end. Each SNP was assigned a p-value based on its strongest joint effect with the other SNPs. We then used the Exploratory Visual Analysis (EVA) method and software to assign a p-value to each gene based on the overabundance of significant SNPs at the α = 0.05 level in the gene. We also used EVA to assign p-values to each GO group based on the overabundance of significant genes at the α = 0.05 level. A GO category was determined to replicate if that category was significant at the α = 0.05 level in both studies. We found two GO categories that replicated in both studies. The first, 'Regulation of Cellular Component Organization and Biogenesis', a GO Biological Process, had p-values of 0.010 and 0.014 in the detection and replication studies, respectively. The second, 'Actin Cytoskeleton', a GO Cellular Component, had p-values of 0.040 and 0.046 in the detection and replication studies, respectively.
Conclusions: Pathway analysis of pairwise genetic associations in two GWAS of sporadic ALS revealed a set of genes involved in cellular component organization and actin cytoskeleton, more specifically, that were not reported by prior GWAS. However, prior biological studies have implicated actin cytoskeleton in ALS and other motor neuron diseases. This study supports the idea that pathway-level analysis of GWAS data may discover important associations not revealed using conventional one-SNP-at-a-time approaches.
Figures
References
-
- ALS Association. http://www.alsa.org/about-als/facts-you-should-know.html.
-
- Cronin S, Berger S, Ding J, Schymick JC, Washecka N, Hernandez DG, Greenway MJ, Bradley DG, Traynor BJ, Hardiman O. A genome-wide association study of sporadic ALS in a homogenous Irish population. Hum Mol Genet. 2008;17(5):768–774. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous