New variants of porcine epidemic diarrhea virus, China, 2011
- PMID: 22840964
- PMCID: PMC3414035
- DOI: 10.3201/eid1808.120002
New variants of porcine epidemic diarrhea virus, China, 2011
Abstract
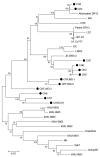
In 2011, porcine epidemic diarrhea virus (PEDV) infection rates rose substantially in vaccinated swine herds. To determine the distribution profile of PEDV outbreak strains, we sequenced the full-length spike gene from samples from 9 farms where animals exhibited severe diarrhea and mortality rates were high. Three new PEDV variants were identified.
Figures

References
-
- Lee DK, Cha SY, Lee C. The N-terminal region of the porcine epidemic diarrhea virus spike protein is important for the receptor binding. Korean Journal of Microbiology and Biotechnology. 2011;39:140–5.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
