A functional framework for interpretation of genetic associations in T1D
- PMID: 22841349
- PMCID: PMC4260931
- DOI: 10.1016/j.coi.2012.07.003
A functional framework for interpretation of genetic associations in T1D
Abstract
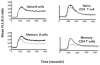
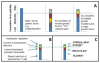
Susceptibility to type 1 diabetes is attributable to genes that link disease progression to distinct steps in immune activation, expansion, and regulation. Recent studies illustrate examples of disease-associated variants that function in multiple cell types and independent pathways, some that impact different steps of a single mechanistic pathway, and some that are functionally interactive for deterministic events in setting thresholds for immune response.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures

References
-
- Robinson DM, Holbeck S, Palmer J, Nepom GT. HLA DQ beta 3.2 identifies subtypes of DR4+ haplotypes permissive for IDDM. Genet Epidemiol. 1989;6:149–154. - PubMed
-
- Nepom GT, Kwok WW. Molecular basis for HLA-DQ associations with IDDM. Diabetes. 1998;47:1177–1184. - PubMed
-
- Erlich H, Valdes AM, Noble J, Carlson JA, Varney M, Concannon P, Mychaleckyj JC, Todd JA, Bonella P, Fear AL, Lavant E, Louey A, Moonsamy P. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2008;57:1084–1092. - PMC - PubMed
-
- Noble JA, Valdes AM, Thomson G, Erlich HA. The HLA class II locus DPB1 can influence susceptibility to type 1 diabetes. Diabetes. 2000;49:121–125. - PubMed
-
- Cucca F, Dudbridge F, Loddo M, Mulargia AP, Lampis R, Angius E, De VS, Koeleman BP, Bain SC, Barnett AH, Gilchrist F, Cordell H, Welsh K, Todd JA. The HLA-DPB1--associated component of the IDDM1 and its relationship to the major loci HLA-DQB1, -DQA1, and -DRB1. Diabetes. 2001;50:1200–1205. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

