The neuroimaging signal is a linear sum of neurally distinct stimulus- and task-related components
- PMID: 22842146
- PMCID: PMC3690535
- DOI: 10.1038/nn.3170
The neuroimaging signal is a linear sum of neurally distinct stimulus- and task-related components
Abstract
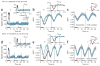
Neuroimaging (for example, functional magnetic resonance imaging) signals are taken as a uniform proxy for local neural activity. By simultaneously recording electrode and neuroimaging (intrinsic optical imaging) signals in alert, task-engaged macaque visual cortex, we recently observed a large anticipatory trial-related neuroimaging signal that was poorly related to local spiking or field potentials. We used these same techniques to study the interactions of this trial-related signal with stimulus-evoked responses over the full range of stimulus intensities, including total darkness. We found that the two signals could be separated, and added linearly over this full range. The stimulus-evoked component was related linearly to local spiking and, consequently, could be used to obtain precise and reliable estimates of local neural activity. The trial-related signal likely has a distinct neural mechanism, however, and failure to account for it properly could lead to substantial errors when estimating local neural spiking from the neuroimaging signal.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


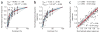


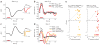
Similar articles
-
Spatial homogeneity and task-synchrony of the trial-related hemodynamic signal.Neuroimage. 2012 Feb 1;59(3):2783-97. doi: 10.1016/j.neuroimage.2011.10.019. Epub 2011 Oct 19. Neuroimage. 2012. PMID: 22036678 Free PMC article.
-
Stimulus-related neuroimaging in task-engaged subjects is best predicted by concurrent spiking.J Neurosci. 2014 Oct 15;34(42):13878-91. doi: 10.1523/JNEUROSCI.1595-14.2014. J Neurosci. 2014. PMID: 25319685 Free PMC article.
-
Anticipatory haemodynamic signals in sensory cortex not predicted by local neuronal activity.Nature. 2009 Jan 22;457(7228):475-9. doi: 10.1038/nature07664. Nature. 2009. PMID: 19158795 Free PMC article.
-
Anticipatory and stimulus-evoked blood oxygenation level-dependent modulations related to spatial attention reflect a common additive signal.J Neurosci. 2009 Aug 26;29(34):10671-82. doi: 10.1523/JNEUROSCI.1141-09.2009. J Neurosci. 2009. PMID: 19710319 Free PMC article.
-
Large-scale visuomotor integration in the cerebral cortex.Cereb Cortex. 2007 Jan;17(1):44-62. doi: 10.1093/cercor/bhj123. Epub 2006 Feb 1. Cereb Cortex. 2007. PMID: 16452643
Cited by
-
Is navigation in virtual reality with FMRI really navigation?J Cogn Neurosci. 2013 Jul;25(7):1008-19. doi: 10.1162/jocn_a_00386. Epub 2013 Mar 14. J Cogn Neurosci. 2013. PMID: 23489142 Free PMC article. Review.
-
Aging drives cerebrovascular network remodeling and functional changes in the mouse brain.bioRxiv [Preprint]. 2023 May 24:2023.05.23.541998. doi: 10.1101/2023.05.23.541998. bioRxiv. 2023. Update in: Nat Commun. 2024 Jul 30;15(1):6398. doi: 10.1038/s41467-024-50559-8. PMID: 37305850 Free PMC article. Updated. Preprint.
-
Single neuron recordings of bilinguals performing in a continuous recognition memory task.PLoS One. 2017 Aug 23;12(8):e0181850. doi: 10.1371/journal.pone.0181850. eCollection 2017. PLoS One. 2017. PMID: 28832639 Free PMC article.
-
Fast hemodynamic responses in the visual cortex of the awake mouse.J Neurosci. 2013 Nov 13;33(46):18343-51. doi: 10.1523/JNEUROSCI.2130-13.2013. J Neurosci. 2013. PMID: 24227743 Free PMC article.
-
Sensory-Evoked Intrinsic Imaging Signals in the Olfactory Bulb Are Independent of Neurovascular Coupling.Cell Rep. 2015 Jul 14;12(2):313-25. doi: 10.1016/j.celrep.2015.06.016. Epub 2015 Jul 2. Cell Rep. 2015. PMID: 26146075 Free PMC article.
References
-
- Heeger DJ, Huk AC, Geisler WS, Albrecht DG. Spikes versus BOLD: what does neuroimaging tell us about neuronal activity? Nat Neurosci. 2000;3:631–633. - PubMed
-
- Rees G, Friston KJ, Koch C. A direct quantitative relationship between the functional properties of human and macaque V5. Nat Neurosci. 2000;3:716–723. - PubMed
-
- Heeger DJ, Ress D. What does fMRI tell us about neural activity? Nat Rev Neurosci. 2002;3:142–151. - PubMed
-
- Friston KJ, Jezzard P, Turner R. Analysis of functional MRI time-series. Hum Brain Mapp. 1994;1:153–171.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

