Tumor-infiltrating DCs suppress nucleic acid-mediated innate immune responses through interactions between the receptor TIM-3 and the alarmin HMGB1
- PMID: 22842346
- PMCID: PMC3622453
- DOI: 10.1038/ni.2376
Tumor-infiltrating DCs suppress nucleic acid-mediated innate immune responses through interactions between the receptor TIM-3 and the alarmin HMGB1
Abstract
The mechanisms by which tumor microenvironments modulate nucleic acid-mediated innate immunity remain unknown. Here we identify the receptor TIM-3 as key in circumventing the stimulatory effects of nucleic acids in tumor immunity. Tumor-associated dendritic cells (DCs) in mouse tumors and patients with cancer had high expression of TIM-3. DC-derived TIM-3 suppressed innate immune responses through the recognition of nucleic acids by Toll-like receptors and cytosolic sensors via a galectin-9-independent mechanism. In contrast, TIM-3 interacted with the alarmin HMGB1 to interfere with the recruitment of nucleic acids into DC endosomes and attenuated the therapeutic efficacy of DNA vaccination and chemotherapy by diminishing the immunogenicity of nucleic acids released from dying tumor cells. Our findings define a mechanism whereby tumor microenvironments suppress antitumor immunity mediated by nucleic acids.
Figures

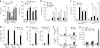
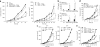
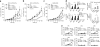
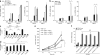


Comment in
-
Tumor immunity times out: TIM-3 and HMGB1.Nat Immunol. 2012 Sep;13(9):808-10. doi: 10.1038/ni.2396. Nat Immunol. 2012. PMID: 22910384 Free PMC article.
-
Immunology: TIM3 suppresses antitumour DCs.Nat Rev Cancer. 2012 Sep;12(9):584. doi: 10.1038/nrc3349. Nat Rev Cancer. 2012. PMID: 22918413 No abstract available.
-
Tumour immunology: TIM3 suppresses antitumour DCs.Nat Rev Immunol. 2012 Sep;12(9):620-1. doi: 10.1038/nri3288. Nat Rev Immunol. 2012. PMID: 22918461 No abstract available.
References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. - PubMed
-
- Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565–1570. - PubMed
-
- Bindea G, Mlecnik B, Fridman WH, Pages F, Galon J. Natural immunity to cancer in humans. Curr. Opin. Immunol. 2010;22:215–222. - PubMed
-
- Dougan M, Dranoff G. Immune therapy for cancer. Annu. Rev. Immunol. 2009;27:83–117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

