A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response
- PMID: 22842901
- PMCID: PMC4691848
- DOI: 10.1038/nature11262
A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response
Abstract
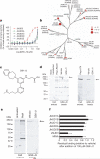
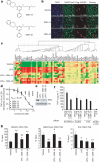
The jumonji (JMJ) family of histone demethylases are Fe2+- and α-ketoglutarate-dependent oxygenases that are essential components of regulatory transcriptional chromatin complexes. These enzymes demethylate lysine residues in histones in a methylation-state and sequence-specific context. Considerable effort has been devoted to gaining a mechanistic understanding of the roles of histone lysine demethylases in eukaryotic transcription, genome integrity and epigenetic inheritance, as well as in development, physiology and disease. However, because of the absence of any selective inhibitors, the relevance of the demethylase activity of JMJ enzymes in regulating cellular responses remains poorly understood. Here we present a structure-guided small-molecule and chemoproteomics approach to elucidating the functional role of the H3K27me3-specific demethylase subfamily (KDM6 subfamily members JMJD3 and UTX). The liganded structures of human and mouse JMJD3 provide novel insight into the specificity determinants for cofactor, substrate and inhibitor recognition by the KDM6 subfamily of demethylases. We exploited these structural features to generate the first small-molecule catalytic site inhibitor that is selective for the H3K27me3-specific JMJ subfamily. We demonstrate that this inhibitor binds in a novel manner and reduces lipopolysaccharide-induced proinflammatory cytokine production by human primary macrophages, a process that depends on both JMJD3 and UTX. Our results resolve the ambiguity associated with the catalytic function of H3K27-specific JMJs in regulating disease-relevant inflammatory responses and provide encouragement for designing small-molecule inhibitors to allow selective pharmacological intervention across the JMJ family.
Figures

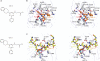
Comment in
-
Structure-based drug design: Opening the door to an epigenetic target.Nat Rev Drug Discov. 2012 Sep;11(9):672. doi: 10.1038/nrd3827. Epub 2012 Aug 20. Nat Rev Drug Discov. 2012. PMID: 22903043 No abstract available.
-
Inhibition of demethylases by GSK-J1/J4.Nature. 2014 Oct 2;514(7520):E1-2. doi: 10.1038/nature13688. Nature. 2014. PMID: 25279926 No abstract available.
-
Kruidenier et al. reply.Nature. 2014 Oct 2;514(7520):E2. doi: 10.1038/nature13689. Nature. 2014. PMID: 25279927 No abstract available.
References
-
- Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 2010;79:155–179. - PubMed
-
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nature Rev. Mol. Cell Biol. 2007;8:307–318. - PubMed
-
- Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nature Rev. Genet. 2007;8:829–833. - PubMed
-
- Trojer P, Reinberg D. Histone lysine demethylases and their impact on epigenetics. Cell. 2006;125:213–217. - PubMed
-
- Loenarz C, Schofield CJ. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem. Sci. 2011;36:7–18. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

