Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis
- PMID: 22842904
- PMCID: PMC3442243
- DOI: 10.1038/nature11376
Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis
Abstract
Ubiquitin modification is mediated by a large family of specificity determining ubiquitin E3 ligases. To facilitate ubiquitin transfer, RING E3 ligases bind both substrate and a ubiquitin E2 conjugating enzyme linked to ubiquitin via a thioester bond, but the mechanism of transfer has remained elusive. Here we report the crystal structure of the dimeric RING domain of rat RNF4 in complex with E2 (UbcH5A) linked by an isopeptide bond to ubiquitin. While the E2 contacts a single protomer of the RING, ubiquitin is folded back onto the E2 by contacts from both RING protomers. The carboxy-terminal tail of ubiquitin is locked into an active site groove on the E2 by an intricate network of interactions, resulting in changes at the E2 active site. This arrangement is primed for catalysis as it can deprotonate the incoming substrate lysine residue and stabilize the consequent tetrahedral transition-state intermediate.
Figures
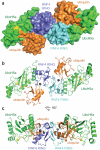

Comment in
-
Structural biology: A protein engagement RING.Nature. 2012 Sep 6;489(7414):43-4. doi: 10.1038/489043a. Nature. 2012. PMID: 22955611 Free PMC article.
Similar articles
-
Structural basis for the RING-catalyzed synthesis of K63-linked ubiquitin chains.Nat Struct Mol Biol. 2015 Aug;22(8):597-602. doi: 10.1038/nsmb.3052. Epub 2015 Jul 6. Nat Struct Mol Biol. 2015. PMID: 26148049 Free PMC article.
-
Mechanism of ubiquitylation by dimeric RING ligase RNF4.Nat Struct Mol Biol. 2011 Aug 21;18(9):1052-9. doi: 10.1038/nsmb.2108. Nat Struct Mol Biol. 2011. PMID: 21857666 Free PMC article.
-
Structure of a HOIP/E2~ubiquitin complex reveals RBR E3 ligase mechanism and regulation.Nature. 2016 Jan 28;529(7587):546-50. doi: 10.1038/nature16511. Epub 2016 Jan 20. Nature. 2016. PMID: 26789245 Free PMC article.
-
Structural Diversity of Ubiquitin E3 Ligase.Molecules. 2021 Nov 4;26(21):6682. doi: 10.3390/molecules26216682. Molecules. 2021. PMID: 34771091 Free PMC article. Review.
-
Structural basis of generic versus specific E2-RING E3 interactions in protein ubiquitination.Protein Sci. 2019 Oct;28(10):1758-1770. doi: 10.1002/pro.3690. Epub 2019 Aug 23. Protein Sci. 2019. PMID: 31340062 Free PMC article. Review.
Cited by
-
How Is the Fidelity of Proteins Ensured in Terms of Both Quality and Quantity at the Endoplasmic Reticulum? Mechanistic Insights into E3 Ubiquitin Ligases.Int J Mol Sci. 2021 Feb 19;22(4):2078. doi: 10.3390/ijms22042078. Int J Mol Sci. 2021. PMID: 33669844 Free PMC article. Review.
-
Ubc13: the Lys63 ubiquitin chain building machine.Oncotarget. 2016 Sep 27;7(39):64471-64504. doi: 10.18632/oncotarget.10948. Oncotarget. 2016. PMID: 27486774 Free PMC article. Review.
-
Function and regulation of SUMO proteases.Nat Rev Mol Cell Biol. 2012 Dec;13(12):755-66. doi: 10.1038/nrm3478. Nat Rev Mol Cell Biol. 2012. PMID: 23175280 Free PMC article. Review.
-
Functional diversification of the NleG effector family in enterohemorrhagic Escherichia coli.Proc Natl Acad Sci U S A. 2018 Oct 2;115(40):10004-10009. doi: 10.1073/pnas.1718350115. Epub 2018 Sep 14. Proc Natl Acad Sci U S A. 2018. PMID: 30217892 Free PMC article.
-
A snapshot of ubiquitin chain elongation: lysine 48-tetra-ubiquitin slows down ubiquitination.J Biol Chem. 2014 Mar 7;289(10):7068-7081. doi: 10.1074/jbc.M113.530576. Epub 2014 Jan 24. J Biol Chem. 2014. PMID: 24464578 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

