Arabidopsis phytochrome a is modularly structured to integrate the multiple features that are required for a highly sensitized phytochrome
- PMID: 22843485
- PMCID: PMC3426125
- DOI: 10.1105/tpc.111.094201
Arabidopsis phytochrome a is modularly structured to integrate the multiple features that are required for a highly sensitized phytochrome
Abstract
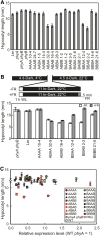
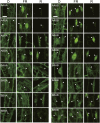
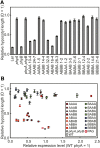
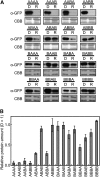
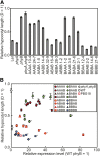
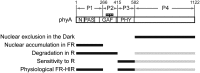
Phytochrome is a red (R)/far-red (FR) light-sensing photoreceptor that regulates various aspects of plant development. Among the members of the phytochrome family, phytochrome A (phyA) exclusively mediates atypical phytochrome responses, such as the FR high irradiance response (FR-HIR), which is elicited under prolonged FR. A proteasome-based degradation pathway rapidly eliminates active Pfr (the FR-absorbing form of phyA) under R. To elucidate the structural basis for the phyA-specific properties, we systematically constructed 16 chimeric phytochromes in which each of four parts of the phytochrome molecule, namely, the N-terminal extension plus the Per/Arnt/Sim domain (N-PAS), the cGMP phosphodiesterase/adenyl cyclase/FhlA domain (GAF), the phytochrome domain (PHY), and the entire C-terminal half, was occupied by either the phyA or phytochrome B sequence. These phytochromes were expressed in transgenic Arabidopsis thaliana to examine their physiological activities. Consequently, the phyA N-PAS sequence was shown to be necessary and sufficient to promote nuclear accumulation under FR, whereas the phyA sequence in PHY was additionally required to exhibit FR-HIR. Furthermore, the phyA sequence in PHY alone substantially increased the light sensitivity to R. In addition, the GAF phyA sequence was important for rapid Pfr degradation. In summary, distinct structural modules, each of which confers different properties to phyA, are assembled on the phyA molecule.
Figures


Similar articles
-
Synergistic and Antagonistic Action of Phytochrome (Phy) A and PhyB during Seedling De-Etiolation in Arabidopsis thaliana.Int J Mol Sci. 2015 May 28;16(6):12199-212. doi: 10.3390/ijms160612199. Int J Mol Sci. 2015. PMID: 26030677 Free PMC article.
-
Arabidopsis phytochrome B promotes SPA1 nuclear accumulation to repress photomorphogenesis under far-red light.Plant Cell. 2013 Jan;25(1):115-33. doi: 10.1105/tpc.112.107086. Epub 2013 Jan 31. Plant Cell. 2013. PMID: 23371951 Free PMC article.
-
Subcellular sites of the signal transduction and degradation of phytochrome A.Plant Cell Physiol. 2010 Oct;51(10):1648-60. doi: 10.1093/pcp/pcq121. Epub 2010 Aug 25. Plant Cell Physiol. 2010. PMID: 20739301
-
Shedding (far-red) light on phytochrome mechanisms and responses in land plants.Plant Sci. 2014 Mar;217-218:36-46. doi: 10.1016/j.plantsci.2013.11.013. Epub 2013 Nov 28. Plant Sci. 2014. PMID: 24467894 Review.
-
Two Distinct Molecular Types of Phytochrome A in Plants: Evidence of Existence and Implications for Functioning.Int J Mol Sci. 2023 May 2;24(9):8139. doi: 10.3390/ijms24098139. Int J Mol Sci. 2023. PMID: 37175844 Free PMC article. Review.
Cited by
-
Mass Spectrometric Analyses Reveal a Central Role for Ubiquitylation in Remodeling the Arabidopsis Proteome during Photomorphogenesis.Mol Plant. 2017 Jun 5;10(6):846-865. doi: 10.1016/j.molp.2017.04.008. Epub 2017 Apr 28. Mol Plant. 2017. PMID: 28461270 Free PMC article.
-
Lysine 206 in Arabidopsis phytochrome A is the major site for ubiquitin-dependent protein degradation.J Biochem. 2016 Feb;159(2):161-9. doi: 10.1093/jb/mvv085. Epub 2015 Aug 26. J Biochem. 2016. PMID: 26314334 Free PMC article.
-
The structure of Arabidopsis phytochrome A reveals topological and functional diversification among the plant photoreceptor isoforms.Nat Plants. 2023 Jul;9(7):1116-1129. doi: 10.1038/s41477-023-01435-8. Epub 2023 Jun 8. Nat Plants. 2023. PMID: 37291396 Free PMC article.
-
Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants.Nat Commun. 2019 Nov 19;10(1):5219. doi: 10.1038/s41467-019-13045-0. Nat Commun. 2019. PMID: 31745087 Free PMC article. Review.
-
From the archives: Where the light goes; flower color, chloroplast transport, and phytochrome A.Plant Cell. 2022 Jul 4;34(7):2570-2571. doi: 10.1093/plcell/koac113. Plant Cell. 2022. PMID: 35474545 Free PMC article. No abstract available.
References
-
- Abe H., Takio K., Titani K., Furuya M. (1989). Amino-terminal amino acid sequences of pea phytochrome II fragments obtained by limited proteolysis. Plant Cell Physiol. 30: 1089–1097
-
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446 - PubMed
-
- Bauer D., Viczián A., Kircher S., Nobis T., Nitschke R., Kunkel T., Panigrahi K.C., Adám E., Fejes E., Schäfer E., Nagy F. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

