Activity of the osmotically regulated yqiHIK promoter from Bacillus subtilis is controlled at a distance
- PMID: 22843846
- PMCID: PMC3457214
- DOI: 10.1128/JB.01041-12
Activity of the osmotically regulated yqiHIK promoter from Bacillus subtilis is controlled at a distance
Abstract
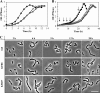
The yqiHIK gene cluster from Bacillus subtilis is predicted to encode an extracellular lipoprotein (YqiH), a secreted N-acetylmuramoyl-L-alanine amidase (YqiI), and a cytoplasmic glycerophosphodiester phosphodiesterase (YqiK). Reverse transcriptase PCR (RT-PCR) analysis showed that the yqiHIK genes are transcribed as an operon. Consistent with the in silico prediction, we found that the purified YqiI protein exhibited hydrolytic activity toward peptidoglycan sacculi. Transcription studies with yqiH-treA reporter fusion strains revealed that the expression of yqiHIK is subjected to finely tuned osmotic control, but enhanced expression occurs only in severely osmotically stressed cells. Primer extension analysis pinpointed the osmotically responsive yqiHIK promoter, and site-directed mutagenesis was employed to assess functionally important sequences required for promoter activity and osmotic control. Promoter variants with constitutive activity were isolated. A deletion analysis of the yqiHIK regulatory region showed that a 53-bp AT-rich DNA segment positioned 180 bp upstream of the -35 sequence is critical for the activity and osmotic regulation of the yqiHIK promoter. Hence, the expression of yqiHIK is subjected to genetic control at a distance. Upon the onset of growth of cells of the B. subtilis wild-type strain in high-salinity medium (1.2 M NaCl), we observed gross morphological deformations of cells that were then reversed to a rod-shaped morphology again when the cells had adjusted to the high-salinity environment. The products of the yqiHIK gene cluster were not critical for reestablishing rod-shaped morphology, but the deletion of this operon yielded a B. subtilis mutant impaired in growth in a defined minimal medium and at high salinity.
Figures






References
-
- Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a Web-based environment for protein structure homology modelling. Bioinformatics 22:195–201 - PubMed
-
- Booth IR, Edwards MD, Black S, Schumann U, Miller S. 2007. Mechanosensitive channels in bacteria: signs of closure? Nat. Rev. Microbiol. 5:431–440 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

