TMBB-DB: a transmembrane β-barrel proteome database
- PMID: 22843985
- PMCID: PMC3463127
- DOI: 10.1093/bioinformatics/bts478
TMBB-DB: a transmembrane β-barrel proteome database
Abstract
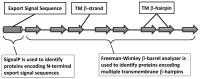
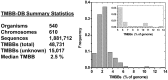
Motivation: We previously reported the development of a highly accurate statistical algorithm for identifying β-barrel outer membrane proteins or transmembrane β-barrels (TMBBs), from genomic sequence data of Gram-negative bacteria (Freeman,T.C. and Wimley,W.C. (2010) Bioinformatics, 26, 1965-1974). We have now applied this identification algorithm to all available Gram-negative bacterial genomes (over 600 chromosomes) and have constructed a publicly available, searchable, up-to-date, database of all proteins in these genomes.
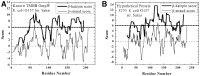
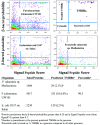
Results: For each protein in the database, there is information on (i) β-barrel membrane protein probability for identification of β-barrels, (ii) β-strand and β-hairpin propensity for structure and topology prediction, (iii) signal sequence score because most TMBBs are secreted through the inner membrane translocon and, thus, have a signal sequence, and (iv) transmembrane α-helix predictions, for reducing false positive predictions. This information is sufficient for the accurate identification of most β-barrel membrane proteins in these genomes. In the database there are nearly 50 000 predicted TMBBs (out of 1.9 million total putative proteins). Of those, more than 15 000 are 'hypothetical' or 'putative' proteins, not previously identified as TMBBs. This wealth of genomic information is not available anywhere else.
Availability: The TMBB genomic database is available at http://beta-barrel.tulane.edu/.
Contact: wwimley@tulane.edu.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

