A benchmark for chromatin binding measurements in live cells
- PMID: 22844090
- PMCID: PMC3424588
- DOI: 10.1093/nar/gks701
A benchmark for chromatin binding measurements in live cells
Abstract
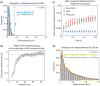
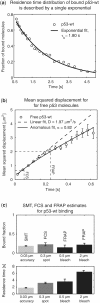
Live-cell measurement of protein binding to chromatin allows probing cellular biochemistry in physiological conditions, which are difficult to mimic in vitro. However, different studies have yielded widely discrepant predictions, and so it remains uncertain how to make the measurements accurately. To establish a benchmark we measured binding of the transcription factor p53 to chromatin by three approaches: fluorescence recovery after photobleaching (FRAP), fluorescence correlation spectroscopy (FCS) and single-molecule tracking (SMT). Using new procedures to analyze the SMT data and to guide the FRAP and FCS analysis, we show how all three approaches yield similar estimates for both the fraction of p53 molecules bound to chromatin (only about 20%) and the residence time of these bound molecules (∼1.8 s). We also apply these procedures to mutants in p53 chromatin binding. Our results support the model that p53 locates specific sites by first binding at sequence-independent sites.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

