Intact p53-dependent responses in miR-34-deficient mice
- PMID: 22844244
- PMCID: PMC3406012
- DOI: 10.1371/journal.pgen.1002797
Intact p53-dependent responses in miR-34-deficient mice
Abstract
MicroRNAs belonging to the miR-34 family have been proposed as critical modulators of the p53 pathway and potential tumor suppressors in human cancers. To formally test these hypotheses, we have generated mice carrying targeted deletion of all three members of this microRNA family. We show that complete inactivation of miR-34 function is compatible with normal development in mice. Surprisingly, p53 function appears to be intact in miR-34-deficient cells and tissues. Although loss of miR-34 expression leads to a slight increase in cellular proliferation in vitro, it does not impair p53-induced cell cycle arrest or apoptosis. Furthermore, in contrast to p53-deficient mice, miR-34-deficient animals do not display increased susceptibility to spontaneous, irradiation-induced, or c-Myc-initiated tumorigenesis. We also show that expression of members of the miR-34 family is particularly high in the testes, lungs, and brains of mice and that it is largely p53-independent in these tissues. These findings indicate that miR-34 plays a redundant function in the p53 pathway and suggest additional p53-independent functions for this family of miRNAs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
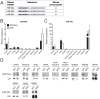


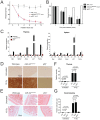
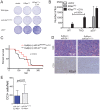
References
-
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. - PubMed
-
- Speidel D. Transcription-independent p53 apoptosis: an alternative route to death. Trends Cell Biol. 2010;20:14–24. - PubMed
-
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. - PubMed
-
- Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–1058. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

