Mg2+ in the major groove modulates B-DNA structure and dynamics
- PMID: 22844516
- PMCID: PMC3402463
- DOI: 10.1371/journal.pone.0041704
Mg2+ in the major groove modulates B-DNA structure and dynamics
Abstract
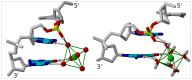
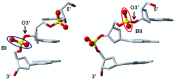
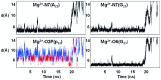
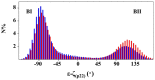
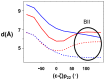
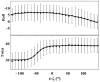
This study investigates the effect of Mg(2+) bound to the DNA major groove on DNA structure and dynamics. The analysis of a comprehensive dataset of B-DNA crystallographic structures shows that divalent cations are preferentially located in the DNA major groove where they interact with successive bases of (A/G)pG and the phosphate group of 5'-CpA or TpG. Based on this knowledge, molecular dynamics simulations were carried out on a DNA oligomer without or with Mg(2+) close to an ApG step. These simulations showed that the hydrated Mg(2+) forms a stable intra-strand cross-link between the two purines in solution. ApG generates an electrostatic potential in the major groove that is particularly attractive for cations; its intrinsic conformation is well-adapted to the formation of water-mediated hydrogen bonds with Mg(2+). The binding of Mg(2+) modulates the behavior of the 5'-neighboring step by increasing the BII (ε-ζ>0°) population of its phosphate group. Additional electrostatic interactions between the 5'-phosphate group and Mg(2+) strengthen both the DNA-cation binding and the BII character of the 5'-step. Cation binding in the major groove may therefore locally influence the DNA conformational landscape, suggesting a possible avenue for better understanding how strong DNA distortions can be stabilized in protein-DNA complexes.
Conflict of interest statement
Figures

Similar articles
-
1 A crystal structures of B-DNA reveal sequence-specific binding and groove-specific bending of DNA by magnesium and calcium.J Mol Biol. 2000 Aug 25;301(4):915-45. doi: 10.1006/jmbi.2000.4012. J Mol Biol. 2000. PMID: 10966796
-
Competition between Na⁺ and Rb⁺ in the minor groove of DNA.Phys Rev E Stat Nonlin Soft Matter Phys. 2012 May;85(5 Pt 1):051913. doi: 10.1103/PhysRevE.85.051913. Epub 2012 May 22. Phys Rev E Stat Nonlin Soft Matter Phys. 2012. PMID: 23004793
-
Conformational rearrangement of 1,2-d(GG) intrastrand cis-diammineplatinum crosslinked DNA is driven by counter-ion penetration within the minor groove of the modified site.J Mol Model. 2017 Sep 14;23(10):278. doi: 10.1007/s00894-017-3445-2. J Mol Model. 2017. PMID: 28913561
-
Cations as hydrogen bond donors: a view of electrostatic interactions in DNA.Annu Rev Biophys Biomol Struct. 2003;32:27-45. doi: 10.1146/annurev.biophys.32.110601.141726. Epub 2003 Feb 14. Annu Rev Biophys Biomol Struct. 2003. PMID: 12598364 Review.
-
A review of the role of the sequence-dependent electrostatic landscape in DNA alkylation patterns.Chem Res Toxicol. 2006 Nov;19(11):1402-14. doi: 10.1021/tx060127n. Chem Res Toxicol. 2006. PMID: 17112226 Free PMC article. Review.
Cited by
-
Bisulfite probing reveals DNA structural intricacies.Nucleic Acids Res. 2023 Apr 24;51(7):3261-3269. doi: 10.1093/nar/gkad115. Nucleic Acids Res. 2023. PMID: 36881756 Free PMC article.
-
The role of assembly parameters on polyplex poly(beta-amino ester) nanoparticle transfections.Biotechnol Bioeng. 2019 May;116(5):1220-1230. doi: 10.1002/bit.26921. Epub 2019 Jan 29. Biotechnol Bioeng. 2019. PMID: 30636286 Free PMC article.
-
A perspective on the molecular simulation of DNA from structural and functional aspects.Chem Sci. 2021 Mar 15;12(15):5390-5409. doi: 10.1039/d0sc05329e. Chem Sci. 2021. PMID: 34168783 Free PMC article. Review.
-
Force regulated dynamics of RPA on a DNA fork.Nucleic Acids Res. 2016 Jul 8;44(12):5837-48. doi: 10.1093/nar/gkw187. Epub 2016 Mar 25. Nucleic Acids Res. 2016. PMID: 27016742 Free PMC article.
-
All-atom crystal simulations of DNA and RNA duplexes.Biochim Biophys Acta. 2015 May;1850(5):1059-1071. doi: 10.1016/j.bbagen.2014.09.018. Epub 2014 Sep 26. Biochim Biophys Acta. 2015. PMID: 25255706 Free PMC article.
References
-
- Egli M. DNA-cation interactions: quo vadis? Chem Biol. 2002;9:277–286. - PubMed
-
- Hud NV, Polak M. DNA-cation interactions: The major and minor grooves are flexible ionophores. Curr Opin Struct Biol. 2001;11:293–301. - PubMed
-
- Subirana JA, Soler-Lopez M. Cations as hydrogen bond donors: a view of electrostatic interactions in DNA. Annu Rev Biophys Biomol Struct. 2003;32:27–45. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

