Identification of a radiosensitivity signature using integrative metaanalysis of published microarray data for NCI-60 cancer cells
- PMID: 22846430
- PMCID: PMC3472294
- DOI: 10.1186/1471-2164-13-348
Identification of a radiosensitivity signature using integrative metaanalysis of published microarray data for NCI-60 cancer cells
Abstract
Background: In the postgenome era, a prediction of response to treatment could lead to better dose selection for patients in radiotherapy. To identify a radiosensitive gene signature and elucidate related signaling pathways, four different microarray experiments were reanalyzed before radiotherapy.
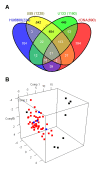
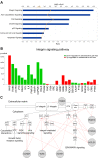
Results: Radiosensitivity profiling data using clonogenic assay and gene expression profiling data from four published microarray platforms applied to NCI-60 cancer cell panel were used. The survival fraction at 2 Gy (SF2, range from 0 to 1) was calculated as a measure of radiosensitivity and a linear regression model was applied to identify genes or a gene set with a correlation between expression and radiosensitivity (SF2). Radiosensitivity signature genes were identified using significant analysis of microarrays (SAM) and gene set analysis was performed using a global test using linear regression model. Using the radiation-related signaling pathway and identified genes, a genetic network was generated. According to SAM, 31 genes were identified as common to all the microarray platforms and therefore a common radiosensitivity signature. In gene set analysis, functions in the cell cycle, DNA replication, and cell junction, including adherence and gap junctions were related to radiosensitivity. The integrin, VEGF, MAPK, p53, JAK-STAT and Wnt signaling pathways were overrepresented in radiosensitivity. Significant genes including ACTN1, CCND1, HCLS1, ITGB5, PFN2, PTPRC, RAB13, and WAS, which are adhesion-related molecules that were identified by both SAM and gene set analysis, and showed interaction in the genetic network with the integrin signaling pathway.
Conclusions: Integration of four different microarray experiments and gene selection using gene set analysis discovered possible target genes and pathways relevant to radiosensitivity. Our results suggested that the identified genes are candidates for radiosensitivity biomarkers and that integrin signaling via adhesion molecules could be a target for radiosensitization.
Figures
References
-
- Begg AC. Predicting response to radiotherapy: evolutions and revolutions. Int J Radiat Biol. 2009;85(10):825–836. - PubMed
-
- Watanabe T, Komuro Y, Kiyomatsu T, Kanazawa T, Kazama Y, Tanaka J, Tanaka T, Yamamoto Y, Shirane M, Muto T. et al. Prediction of sensitivity of rectal cancer cells in response to preoperative radiotherapy by DNA microarray analysis of gene expression profiles. Cancer Res. 2006;66(7):3370–3374. doi: 10.1158/0008-5472.CAN-05-3834. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

