The RosR transcription factor is required for gene expression dynamics in response to extreme oxidative stress in a hypersaline-adapted archaeon
- PMID: 22846541
- PMCID: PMC3443676
- DOI: 10.1186/1471-2164-13-351
The RosR transcription factor is required for gene expression dynamics in response to extreme oxidative stress in a hypersaline-adapted archaeon
Abstract
Background: Previous work has shown that the hypersaline-adapted archaeon, Halobacterium salinarum NRC-1, is highly resistant to oxidative stress caused by exposure to hydrogen peroxide, UV, and gamma radiation. Dynamic alteration of the gene regulatory network (GRN) has been implicated in such resistance. However, the molecular functions of transcription regulatory proteins involved in this response remain unknown.
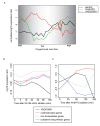
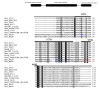
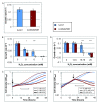
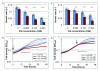
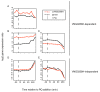
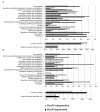
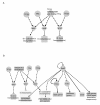
Results: Here we have reanalyzed several existing GRN and systems biology datasets for H. salinarum to identify and characterize a novel winged helix-turn-helix transcription factor, VNG0258H, as a regulator required for reactive oxygen species resistance in this organism. This protein appears to be unique to the haloarchaea at the primary sequence level. High throughput quantitative growth assays in a deletion mutant strain implicate VNG0258H in extreme oxidative stress resistance. According to time course gene expression analyses, this transcription factor is required for the appropriate dynamic response of nearly 300 genes to reactive oxygen species damage from paraquat and hydrogen peroxide. These genes are predicted to function in repair of oxidative damage to proteins and DNA. In vivo DNA binding assays demonstrate that VNG0258H binds DNA to mediate gene regulation.
Conclusions: Together these results suggest that VNG0258H is a novel archaeal transcription factor that regulates gene expression to enable adaptation to the extremely oxidative, hypersaline niche of H. salinarum. We have therefore renamed VNG0258H as RosR, for reactive oxygen species regulator.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

