Structural basis of the allosteric inhibitor interaction on the HIV-1 reverse transcriptase RNase H domain
- PMID: 22846652
- PMCID: PMC3465473
- DOI: 10.1111/cbdd.12010
Structural basis of the allosteric inhibitor interaction on the HIV-1 reverse transcriptase RNase H domain
Abstract
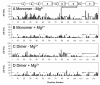
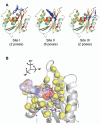
HIV-1 reverse transcriptase (RT) has been an attractive target for the development of antiretroviral agents. Although this enzyme is bi-functional, having both DNA polymerase and ribonuclease H (RNH) activities, there is no clinically approved inhibitor of the RNH activity. Here, we characterize the structural basis and molecular interaction of an allosteric site inhibitor, BHMP07, with the wild-type (WT) RNH fragment. Solution NMR experiments for inhibitor titration on WT RNH showed relatively wide chemical shift perturbations, suggesting a long-range conformational effect on the inhibitor interaction. Comparisons of the inhibitor-induced NMR chemical shift changes of RNH with those of RNH dimer, in the presence and absence of Mg(2+) , were performed to determine and verify the interaction site. The NMR results, with assistance of molecular docking, indicate that BHMP07 preferentially binds to a site that is located between the RNH active site and the region encompassing helices B and D (the 'substrate-handle region'). The interaction site is consistent with the previous proposed site, identified using a chimeric RNH (p15-EC) [Gong et al. (2011) Chem Biol Drug Des 77, 39-47], but with slight differences that reflect the characteristics of the amino acid sequences in p15-EC compared to the WT RNH.
© 2012 John Wiley & Sons A/S.
Figures


Similar articles
-
Interaction of HIV-1 reverse transcriptase ribonuclease H with an acylhydrazone inhibitor.Chem Biol Drug Des. 2011 Jan;77(1):39-47. doi: 10.1111/j.1747-0285.2010.01052.x. Epub 2010 Nov 29. Chem Biol Drug Des. 2011. PMID: 21114787 Free PMC article.
-
A 2-Hydroxyisoquinoline-1,3-Dione Active-Site RNase H Inhibitor Binds in Multiple Modes to HIV-1 Reverse Transcriptase.Antimicrob Agents Chemother. 2017 Sep 22;61(10):e01351-17. doi: 10.1128/AAC.01351-17. Print 2017 Oct. Antimicrob Agents Chemother. 2017. PMID: 28760905 Free PMC article.
-
HIV-1 reverse transcriptase structure with RNase H inhibitor dihydroxy benzoyl naphthyl hydrazone bound at a novel site.ACS Chem Biol. 2006 Dec 20;1(11):702-12. doi: 10.1021/cb600303y. ACS Chem Biol. 2006. PMID: 17184135 Free PMC article.
-
Past and future. Current drugs targeting HIV-1 integrase and reverse transcriptase-associated ribonuclease H activity: single and dual active site inhibitors.Antivir Chem Chemother. 2014 Jan 29;23(4):129-44. doi: 10.3851/IMP2690. Antivir Chem Chemother. 2014. PMID: 24150519 Review.
-
Novel HIV-1 reverse transcriptase inhibitors.Virus Res. 2008 Jun;134(1-2):171-85. doi: 10.1016/j.virusres.2008.01.003. Epub 2008 Mar 4. Virus Res. 2008. PMID: 18308412 Review.
Cited by
-
Inhibitors of HIV-1 Reverse Transcriptase-Associated Ribonuclease H Activity.Biology (Basel). 2012 Oct 19;1(3):521-41. doi: 10.3390/biology1030521. Biology (Basel). 2012. PMID: 23599900 Free PMC article.
-
Ribonuclease H/DNA Polymerase HIV-1 Reverse Transcriptase Dual Inhibitor: Mechanistic Studies on the Allosteric Mode of Action of Isatin-Based Compound RMNC6.PLoS One. 2016 Jan 22;11(1):e0147225. doi: 10.1371/journal.pone.0147225. eCollection 2016. PLoS One. 2016. PMID: 26800261 Free PMC article.
-
Inhibition of foamy virus reverse transcriptase by human immunodeficiency virus type 1 RNase H inhibitors.Antimicrob Agents Chemother. 2014 Jul;58(7):4086-93. doi: 10.1128/AAC.00056-14. Epub 2014 May 5. Antimicrob Agents Chemother. 2014. PMID: 24798282 Free PMC article.
-
Discovery of a small-molecule binder of the oncoprotein gankyrin that modulates gankyrin activity in the cell.Sci Rep. 2016 Apr 5;6:23732. doi: 10.1038/srep23732. Sci Rep. 2016. PMID: 27046077 Free PMC article.
-
Free Energy-Based Virtual Screening and Optimization of RNase H Inhibitors of HIV-1 Reverse Transcriptase.ACS Omega. 2016 Sep 30;1(3):435-447. doi: 10.1021/acsomega.6b00123. Epub 2016 Sep 21. ACS Omega. 2016. PMID: 27713931 Free PMC article.
References
-
- Sarafianos SG, Das K, Hughes SH, Arnold E. Taking aim at a moving target: designing drugs to inhibit drug-resistant HIV-1 reverse transcriptases. Curr Opin Struct Biol. 2004;14:716–30. - PubMed
-
- De Clercq E. Non-nucleoside reverse transcriptase inhibitors (NNRTIs): past, present, and future. Chem Biodivers. 2004;1:44–64. - PubMed
-
- Basavapathruni A, Anderson KS. Reverse transcription of the HIV-1 pandemic. FASEB J. 2007;21:3795–808. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

