Storage conditions of intestinal microbiota matter in metagenomic analysis
- PMID: 22846661
- PMCID: PMC3489833
- DOI: 10.1186/1471-2180-12-158
Storage conditions of intestinal microbiota matter in metagenomic analysis
Abstract
Background: The structure and function of human gut microbiota is currently inferred from metagenomic and metatranscriptomic analyses. Recovery of intact DNA and RNA is therefore a critical step in these studies. Here, we evaluated how different storage conditions of fecal samples affect the quality of extracted nucleic acids and the stability of their microbial communities.
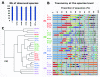
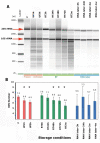
Results: We assessed the quality of genomic DNA and total RNA by microcapillary electrophoresis and analyzed the bacterial community structure by pyrosequencing the 16S rRNA gene. DNA and RNA started to fragment when samples were kept at room temperature for more than 24 h. The use of RNAse inhibitors diminished RNA degradation but this protection was not consistent among individuals. DNA and RNA degradation also occurred when frozen samples were defrosted for a short period (1 h) before nucleic acid extraction. The same conditions that affected DNA and RNA integrity also altered the relative abundance of most taxa in the bacterial community analysis. In this case, intra-individual variability of microbial diversity was larger than inter-individual one.
Conclusions: Though this preliminary work explored a very limited number of parameters, the results suggest that storage conditions of fecal samples affect the integrity of DNA and RNA and the composition of their microbial community. For optimal preservation, stool samples should be kept at room temperature and brought at the laboratory within 24 h after collection or be stored immediately at -20°C in a home freezer and transported afterwards in a freezer pack to ensure that they do not defrost at any time. Mixing the samples with RNAse inhibitors outside the laboratory is not recommended since proper homogenization of the stool is difficult to monitor.
Figures
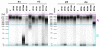
Similar articles
-
Comparison of stool versus rectal swab samples and storage conditions on bacterial community profiles.BMC Microbiol. 2017 Mar 31;17(1):78. doi: 10.1186/s12866-017-0983-9. BMC Microbiol. 2017. PMID: 28359329 Free PMC article.
-
Characterization of the fecal microbiota using high-throughput sequencing reveals a stable microbial community during storage.PLoS One. 2012;7(10):e46953. doi: 10.1371/journal.pone.0046953. Epub 2012 Oct 5. PLoS One. 2012. PMID: 23071673 Free PMC article.
-
Comparison of methods for fecal microbiome biospecimen collection.BMC Microbiol. 2014 Apr 23;14:103. doi: 10.1186/1471-2180-14-103. BMC Microbiol. 2014. PMID: 24758293 Free PMC article.
-
Effects of Stool Sample Preservation Methods on Gut Microbiota Biodiversity: New Original Data and Systematic Review with Meta-Analysis.Microbiol Spectr. 2023 Jun 15;11(3):e0429722. doi: 10.1128/spectrum.04297-22. Epub 2023 Apr 24. Microbiol Spectr. 2023. PMID: 37093040 Free PMC article.
-
Metagenomics of human microbiome: beyond 16s rDNA.Clin Microbiol Infect. 2012 Jul;18 Suppl 4:47-9. doi: 10.1111/j.1469-0691.2012.03865.x. Clin Microbiol Infect. 2012. PMID: 22647049 Review.
Cited by
-
The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects.PLoS One. 2015 May 29;10(5):e0126685. doi: 10.1371/journal.pone.0126685. eCollection 2015. PLoS One. 2015. PMID: 26024217 Free PMC article.
-
Consumption of a Bifidobacterium bifidum Strain for 4 Weeks Modulates Dominant Intestinal Bacterial Taxa and Fecal Butyrate in Healthy Adults.Appl Environ Microbiol. 2016 Sep 16;82(19):5850-9. doi: 10.1128/AEM.01753-16. Print 2016 Oct 1. Appl Environ Microbiol. 2016. PMID: 27451450 Free PMC article. Clinical Trial.
-
A robust ambient temperature collection and stabilization strategy: Enabling worldwide functional studies of the human microbiome.Sci Rep. 2016 Aug 25;6:31731. doi: 10.1038/srep31731. Sci Rep. 2016. PMID: 27558918 Free PMC article.
-
Gut bless you: The microbiota-gut-brain axis in irritable bowel syndrome.World J Gastroenterol. 2022 Jan 28;28(4):412-431. doi: 10.3748/wjg.v28.i4.412. World J Gastroenterol. 2022. PMID: 35125827 Free PMC article. Review.
-
The impact of storage buffer and storage conditions on fecal samples for bacteriophage infectivity and metavirome analyses.Microbiome. 2023 Aug 28;11(1):193. doi: 10.1186/s40168-023-01632-9. Microbiome. 2023. PMID: 37635262 Free PMC article.
References
-
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P. et al.A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. - DOI - PMC - PubMed
-
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S. et al.Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. - DOI - PMC - PubMed
-
- Dolfing J, Vos A, Bloem J, Ehlert PA, Naumova NB, Kuikman PJ. Microbial diversity in archived soils. Science. 2004;306(5697):813. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

