Virulence factors identified by Cryptococcus neoformans mutant screen differentially modulate lung immune responses and brain dissemination
- PMID: 22846723
- PMCID: PMC3463625
- DOI: 10.1016/j.ajpath.2012.06.012
Virulence factors identified by Cryptococcus neoformans mutant screen differentially modulate lung immune responses and brain dissemination
Abstract
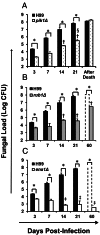
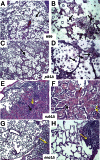
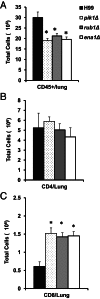
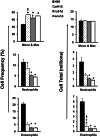
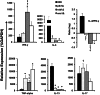
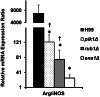
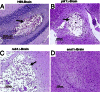
Deletions of cryptococcal PIK1, RUB1, and ENA1 genes independently rendered defects in yeast survival in human CSF and within macrophages. We evaluated virulence potential of these genes by comparing wild-type Cryptococcus neoformans strain H99 with deletant and complement strains in a BALB/c mouse model of pulmonary infection. Survival of infected mice; pulmonary cryptococcal growth and pathology; immunological parameters; dissemination kinetics; and CNS pathology were examined. Deletion of each PIK1, RUB1, and ENA1 differentially reduced pulmonary growth and dissemination rates of C. neoformans and extended mice survival. Furthermore, pik1Δ induced similar pathologies to H99, however, with significantly delayed onset; rub1Δ was more efficiently contained within pulmonary macrophages and was further delayed in causing CNS dissemination/pathology; whereas ena1Δ was progressively eliminated from the lungs and did not induce pathological lesions or disseminate into the CNS. The diminished virulence of mutant strains was associated with differential modulation of pulmonary immune responses, including changes in leukocyte subsets, cytokine responses, and macrophage activation status. Compared to H99 infection, mutants induced more hallmarks of a protective Th1 immune response, rather than Th2, and more classical, rather than alternative, macrophage activation. The magnitude of immunological effects precisely corresponded to the level of virulence displayed by each strain. Thus, cryptococcal PIK1, RUB1, and ENA1 differentially contribute to cryptococcal virulence, in correlation with their differential capacity to modulate immune responses.
Copyright © 2012 American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Huffnagle G.B., Lipscomb M.F., Lovchik J.A., Hoag K.A., Street N.E. The role of CD4+ and CD8+ T cells in the protective inflammatory response to a pulmonary cryptococcal infection. J Leukoc Biol. 1994;55:35–42. - PubMed
-
- Kawakami K., Koguchi Y., Qureshi M.H., Yara S., Kinjo Y., Miyazato A., Nishizawa A., Nariuchi H., Saito A. Circulating soluble CD4 directly prevents host resistance and delayed-type hypersensitivity response to Cryptococcus neoformans in mice. Microbiol Immunol. 2000;44:1033–1041. - PubMed
-
- Traynor T.R., Kuziel W.A., Toews G.B., Huffnagle G.B. CCR2 expression determines T1 versus T2 polarization during pulmonary Cryptococcus neoformans infection. J Immunol. 2000;164:2021–2027. - PubMed
-
- Jain A.V., Zhang Y., Fields W.B., McNamara D.A., Choe M.Y., Chen G.H., Erb-Downward J., Osterholzer J.J., Toews G.B., Huffnagle G.B., Olszewski M.A. Th2 but not Th1 immune bias results in altered lung functions in a murine model of pulmonary Cryptococcus neoformans infection. Infect Immun. 2009;77:5389–5399. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

