Identification of a high affinity nucleocapsid protein binding element from the bovine leukemia virus genome
- PMID: 22846919
- PMCID: PMC3514405
- DOI: 10.1016/j.virusres.2012.07.020
Identification of a high affinity nucleocapsid protein binding element from the bovine leukemia virus genome
Abstract
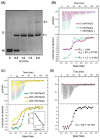
Retroviral genome recognition is mediated by interactions between the nucleocapsid (NC) domain of the virally encoded Gag polyprotein and cognate RNA packaging elements that, for most retroviruses, appear to reside primarily within the 5'-untranslated region (5'-UTR) of the genome. Recent studies suggest that a major packaging determinant of bovine leukemia virus (BLV), a member of the human T-cell leukemia virus (HTLV)/BLV family and a non-primate animal model for HTLV-induced leukemogenesis, resides within the gag open reading frame. We have prepared and purified the recombinant BLV NC protein and conducted electrophoretic mobility shift and isothermal titration calorimetry studies with RNA fragments corresponding to these proposed packaging elements. The gag-derived RNAs did not exhibit significant affinity for NC, suggesting an alternate role in packaging. However, an 83-nucleotide fragment of the 5'-UTR that resides just upstream of the gag start codon binds NC stoichiometrically and with high affinity (K(d)=136±21 nM). These nucleotides were predicted to form tandem hairpin structures, and studies with smaller fragments indicate that the NC binding site resides exclusively within the distal hairpin (residues G369-U399, K(d)=67±8 nM at physiological ionic strength). Unlike all other structurally characterized retroviral NC binding RNAs, this fragment is not expected to contain exposed guanosines, suggesting that RNA binding may be mediated by a previously uncharacterized mechanism.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures



Similar articles
-
Bovine leukemia virus nucleocapsid protein is an efficient nucleic acid chaperone.Biochem Biophys Res Commun. 2015 Mar 13;458(3):687-692. doi: 10.1016/j.bbrc.2015.02.025. Epub 2015 Feb 14. Biochem Biophys Res Commun. 2015. PMID: 25686502
-
Unpaired Guanosines in the 5' Untranslated Region of HIV-1 RNA Act Synergistically To Mediate Genome Packaging.J Virol. 2020 Oct 14;94(21):e00439-20. doi: 10.1128/JVI.00439-20. Print 2020 Oct 14. J Virol. 2020. PMID: 32796062 Free PMC article.
-
Identification of a high affinity nucleocapsid protein binding element within the Moloney murine leukemia virus Psi-RNA packaging signal: implications for genome recognition.J Mol Biol. 2001 Nov 23;314(2):217-32. doi: 10.1006/jmbi.2001.5139. J Mol Biol. 2001. PMID: 11718556
-
Animal models of bovine leukemia virus and human T-lymphotrophic virus type-1: insights in transmission and pathogenesis.Annu Rev Anim Biosci. 2014 Feb;2:189-208. doi: 10.1146/annurev-animal-022513-114117. Epub 2013 Dec 13. Annu Rev Anim Biosci. 2014. PMID: 25384140 Review.
-
Epidemiology and genetic diversity of bovine leukemia virus.Virol J. 2017 Nov 2;14(1):209. doi: 10.1186/s12985-017-0876-4. Virol J. 2017. PMID: 29096657 Free PMC article. Review.
Cited by
-
Human T-cell leukemia virus type 1 Gag domains have distinct RNA-binding specificities with implications for RNA packaging and dimerization.J Biol Chem. 2018 Oct 19;293(42):16261-16276. doi: 10.1074/jbc.RA118.005531. Epub 2018 Sep 14. J Biol Chem. 2018. PMID: 30217825 Free PMC article.
-
Identification of the initial nucleocapsid recognition element in the HIV-1 RNA packaging signal.Proc Natl Acad Sci U S A. 2020 Jul 28;117(30):17737-17746. doi: 10.1073/pnas.2008519117. Epub 2020 Jul 9. Proc Natl Acad Sci U S A. 2020. PMID: 32647061 Free PMC article.
References
-
- Amarasinghe GK, De Guzman RN, Turner RB, Chancellor K, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the ψ-RNA packaging signal. J Mol Biol. 2000;301:491–511. - PubMed
-
- Amarasinghe GK, Zhou J, Miskimon M, Chancellor KJ, McDonald JA, Matthews AG, Miller RA, Rouse MD, Summers MF. Stem-loop SL4 of the HIV-1 ψ-RNA packaging signal exhibits weak affinity for the nucleocapsid protein. Structural studies and implications for genome recognition. J Mol Biol. 2001;314:961–969. - PubMed
-
- Berkhout B. Prog Nucl Acid Res and Mol Biol. Vol. 54. Academic Press, Inc; 1996. Structure and function of the human immunodeficiency virus leader RNA; pp. 1–34. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

