Nuclear-to-cytoplasmic relocalization of the proliferating cell nuclear antigen (PCNA) during differentiation involves a chromosome region maintenance 1 (CRM1)-dependent export and is a prerequisite for PCNA antiapoptotic activity in mature neutrophils
- PMID: 22846997
- PMCID: PMC3460476
- DOI: 10.1074/jbc.M112.367839
Nuclear-to-cytoplasmic relocalization of the proliferating cell nuclear antigen (PCNA) during differentiation involves a chromosome region maintenance 1 (CRM1)-dependent export and is a prerequisite for PCNA antiapoptotic activity in mature neutrophils
Abstract
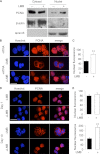
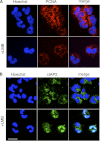
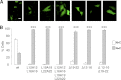
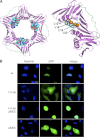
Neutrophils are deprived of proliferative capacity and have a tightly controlled lifespan to avoid their persistence at the site of injury. We have recently described that the proliferating cell nuclear antigen (PCNA), a nuclear factor involved in DNA replication and repair of proliferating cells, is a key regulator of neutrophil survival. In neutrophils, PCNA was localized exclusively in the cytoplasm due to its nuclear-to-cytoplasmic relocalization during granulocytic differentiation. We showed here that leptomycin B, an inhibitor of the chromosome region maintenance 1 (CRM1) exportin, inhibited PCNA relocalization during granulocytic differentiation of HL-60 and NB4 promyelocytic cell lines and of human CD34(+) primary cells. Using enhanced green fluorescent protein fusion constructs, we have demonstrated that PCNA relocalization involved a nuclear export signal (NES) located from Ile-11 to Ile-23 in the PCNA sequence. However, this NES, located at the inner face of the PCNA trimer, was not functional in wild-type PCNA, but instead, was fully active and leptomycin B-sensitive in the monomeric PCNAY114A mutant. To test whether a defect in PCNA cytoplasmic relocalization would affect its antiapoptotic activity in mature neutrophils, a chimeric PCNA fused with the SV40 nuclear localization sequence (NLS) was generated to preclude its cytoplasmic localization. As expected, neutrophil-differentiated PLB985 cells expressing ectopic SV40NLS-PCNA had an increased nuclear PCNA as compared with cells expressing wild-type PCNA. Accordingly, the nuclear PCNA mutant did not show any antiapoptotic activity as compared with wild-type PCNA. Nuclear-to-cytoplasmic relocalization that occurred during myeloid differentiation is essential for PCNA antiapoptotic activity in mature neutrophils and is dependent on the newly identified monomerization-dependent PCNA NES.
Figures



Similar articles
-
Characterization of cytosolic proliferating cell nuclear antigen (PCNA) in neutrophils: antiapoptotic role of the monomer.J Leukoc Biol. 2013 Oct;94(4):723-31. doi: 10.1189/jlb.1212637. Epub 2013 Jul 3. J Leukoc Biol. 2013. PMID: 23825390
-
Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1.Exp Cell Res. 1998 Aug 1;242(2):540-7. doi: 10.1006/excr.1998.4136. Exp Cell Res. 1998. PMID: 9683540
-
CRM1 mediates the nuclear export of YTHDF2.Biochem Biophys Res Commun. 2025 Jun 30;767:151899. doi: 10.1016/j.bbrc.2025.151899. Epub 2025 Apr 25. Biochem Biophys Res Commun. 2025. PMID: 40315567
-
Inhibition of CRM1-dependent nuclear export sensitizes malignant cells to cytotoxic and targeted agents.Semin Cancer Biol. 2014 Aug;27:62-73. doi: 10.1016/j.semcancer.2014.03.001. Epub 2014 Mar 12. Semin Cancer Biol. 2014. PMID: 24631834 Free PMC article. Review.
-
Dynamics of galectin-3 in the nucleus and cytoplasm.Biochim Biophys Acta. 2010 Feb;1800(2):181-9. doi: 10.1016/j.bbagen.2009.07.005. Epub 2009 Jul 16. Biochim Biophys Acta. 2010. PMID: 19616076 Free PMC article. Review.
Cited by
-
Proliferating Cell Nuclear Antigen Suppresses RNA Replication of Bamboo Mosaic Virus through an Interaction with the Viral Genome.J Virol. 2019 Oct 29;93(22):e00961-19. doi: 10.1128/JVI.00961-19. Print 2019 Nov 15. J Virol. 2019. PMID: 31511381 Free PMC article.
-
CCR2 Chemokine Receptors Enhance Growth and Cell-Cycle Progression of Breast Cancer Cells through SRC and PKC Activation.Mol Cancer Res. 2019 Feb;17(2):604-617. doi: 10.1158/1541-7786.MCR-18-0750. Epub 2018 Nov 16. Mol Cancer Res. 2019. PMID: 30446625 Free PMC article.
-
Micropeptide ASAP encoded by LINC00467 promotes colorectal cancer progression by directly modulating ATP synthase activity.J Clin Invest. 2021 Nov 15;131(22):e152911. doi: 10.1172/JCI152911. J Clin Invest. 2021. PMID: 34591791 Free PMC article.
-
mAb14, a Monoclonal Antibody against Cell Surface PCNA: A Potential Tool for Sezary Syndrome Diagnosis and Targeted Immunotherapy.Cancers (Basel). 2023 Sep 4;15(17):4421. doi: 10.3390/cancers15174421. Cancers (Basel). 2023. PMID: 37686697 Free PMC article.
-
Immunohistochemical Detection of Iron-Related Proteins in Sertoli Cell-Only Patterns in Canine Testicular Lesions.Animals (Basel). 2025 May 9;15(10):1377. doi: 10.3390/ani15101377. Animals (Basel). 2025. PMID: 40427255 Free PMC article.
References
-
- Borregaard N. (2010) Neutrophils, from marrow to microbes. Immunity 33, 657–670 - PubMed
-
- Witko-Sarsat V., Rieu P., Descamps-Latscha B., Lesavre P., Halbwachs-Mecarelli L. (2000) Neutrophils: molecules, functions, and pathophysiological aspects. Lab. Invest. 80, 617–653 - PubMed
-
- Simon H. U. (2003) Neutrophil apoptosis pathways and their modifications in inflammation. Immunol. Rev. 193, 101–110 - PubMed
-
- Rossi A. G., Sawatzky D. A., Walker A., Ward C., Sheldrake T. A., Riley N. A., Caldicott A., Martinez-Losa M., Walker T. R., Duffin R., Gray M., Crescenzi E., Martin M. C., Brady H. J., Savill J. S., Dransfield I., Haslett C. (2006) Cyclin-dependent kinase inhibitors enhance the resolution of inflammation by promoting inflammatory cell apoptosis. Nat. Med. 12, 1056–1064 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

