Global expression profiling of globose basal cells and neurogenic progression within the olfactory epithelium
- PMID: 22847514
- PMCID: PMC3887395
- DOI: 10.1002/cne.23204
Global expression profiling of globose basal cells and neurogenic progression within the olfactory epithelium
Abstract
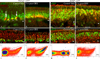
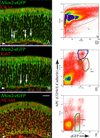
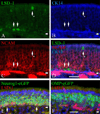
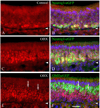
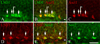
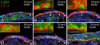
Ongoing, lifelong neurogenesis maintains the neuronal population of the olfactory epithelium in the face of piecemeal neuronal turnover and restores it following wholesale loss. The molecular phenotypes corresponding to different stages along the progression from multipotent globose basal cell (GBC) progenitor to differentiated olfactory sensory neuron are poorly characterized. We used the transgenic expression of enhanced green fluorescent protein (eGFP) and cell surface markers to FACS-isolate ΔSox2-eGFP(+) GBCs, Neurog1-eGFP(+) GBCs and immature neurons, and ΔOMP-eGFP(+) mature neurons from normal adult mice. In addition, the latter two populations were also collected 3 weeks after olfactory bulb ablation, a lesion that results in persistently elevated neurogenesis. Global profiling of mRNA from the populations indicates that all stages of neurogenesis share a cohort of >2,100 genes that are upregulated compared to sustentacular cells. A further cohort of >1,200 genes are specifically upregulated in GBCs as compared to sustentacular cells and differentiated neurons. The increased rate of neurogenesis caused by olfactory bulbectomy had little effect on the transcriptional profile of the Neurog1-eGFP(+) population. In contrast, the abbreviated lifespan of ΔOMP-eGFP(+) neurons born in the absence of the bulb correlated with substantial differences in gene expression as compared to the mature neurons of the normal epithelium. Detailed examination of the specific genes upregulated in the different progenitor populations revealed that the chromatin modifying complex proteins LSD1 and coREST were expressed sequentially in upstream ΔSox2-eGFP(+) GBCs and Neurog1-eGFP(+) GBCs/immature neurons. The expression patterns of these proteins are dynamically regulated after activation of the epithelium by methyl bromide lesion.
Copyright © 2012 Wiley Periodicals, Inc.
Conflict of interest statement
The authors declare that no conflict of interest exists.
Figures


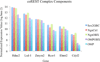

Similar articles
-
The Neuroregenerative Capacity of Olfactory Stem Cells Is Not Limitless: Implications for Aging.J Neurosci. 2018 Aug 1;38(31):6806-6824. doi: 10.1523/JNEUROSCI.3261-17.2018. Epub 2018 Jun 22. J Neurosci. 2018. PMID: 29934351 Free PMC article.
-
Multipotency of purified, transplanted globose basal cells in olfactory epithelium.J Comp Neurol. 2004 Feb 16;469(4):457-74. doi: 10.1002/cne.11031. J Comp Neurol. 2004. PMID: 14755529
-
Wnt-responsive Lgr5⁺ globose basal cells function as multipotent olfactory epithelium progenitor cells.J Neurosci. 2014 Jun 11;34(24):8268-76. doi: 10.1523/JNEUROSCI.0240-14.2014. J Neurosci. 2014. PMID: 24920630 Free PMC article.
-
Stem and progenitor cells of the mammalian olfactory epithelium: Taking poietic license.J Comp Neurol. 2017 Mar 1;525(4):1034-1054. doi: 10.1002/cne.24105. Epub 2016 Sep 27. J Comp Neurol. 2017. PMID: 27560601 Free PMC article. Review.
-
Identification and molecular regulation of neural stem cells in the olfactory epithelium.Exp Cell Res. 2005 Jun 10;306(2):309-16. doi: 10.1016/j.yexcr.2005.03.027. Epub 2005 Apr 21. Exp Cell Res. 2005. PMID: 15925585 Review.
Cited by
-
An epigenetic trap stabilizes singular olfactory receptor expression.Cell. 2013 Jul 18;154(2):325-36. doi: 10.1016/j.cell.2013.06.039. Cell. 2013. PMID: 23870122 Free PMC article.
-
Lysine-specific demethylase-1 (LSD1) is compartmentalized at nuclear chromocenters in early post-mitotic cells of the olfactory sensory neuronal lineage.Mol Cell Neurosci. 2016 Jul;74:58-70. doi: 10.1016/j.mcn.2016.03.001. Epub 2016 Mar 4. Mol Cell Neurosci. 2016. PMID: 26947098 Free PMC article.
-
Olfactory receptor coding sequences cause silencing of episomal constructs in multiple cell lines.Mol Cell Neurosci. 2021 Dec;117:103681. doi: 10.1016/j.mcn.2021.103681. Epub 2021 Nov 4. Mol Cell Neurosci. 2021. PMID: 34742908 Free PMC article.
-
Expression of the SARS-CoV-2 Entry Proteins, ACE2 and TMPRSS2, in Cells of the Olfactory Epithelium: Identification of Cell Types and Trends with Age.ACS Chem Neurosci. 2020 Jun 3;11(11):1555-1562. doi: 10.1021/acschemneuro.0c00210. Epub 2020 May 19. ACS Chem Neurosci. 2020. PMID: 32379417 Free PMC article.
-
A lifetime of neurogenesis in the olfactory system.Front Neurosci. 2014 Jun 26;8:182. doi: 10.3389/fnins.2014.00182. eCollection 2014. Front Neurosci. 2014. PMID: 25018692 Free PMC article. Review.
References
-
- Agustsson T, Nilsson M, Henriksson G, Arnelo U, Juto JE, Lundell L. Treatment of postoperative esophagorespiratory fistulas with dual self-expanding metal stents. World J Surg. 2009;33:1224–1228. - PubMed
-
- Aubert J, Stavridis MP, Tweedie S, O’Reilly M, Vierlinger K, Li M, Ghazal P, Pratt T, Mason JO, Roy D, Smith A. Screening for mammalian neural genes via fluorescence-activated cell sorter purification of neural precursors from Sox1-gfp knock-in mice. Proc Natl Acad Sci U S A. 2003;100(Suppl 1):11836–11841. - PMC - PubMed
-
- Baker H, Grillo M, Margolis FL. Biochemical and immunocytochemical characterization of olfactory marker protein in the rodent central nervous system. J Comp Neurol. 1989;285:246–261. - PubMed
-
- Barraud P, He X, Zhao C, Ibanez C, Raha-Chowdhury R, Cald-well MA, Franklin RJ. Contrasting effects of basic fibroblast growth factor and epidermal growth factor on mouse neonatal olfactory mucosa cells. Eur J Neurosci. 2007;26:3345–3357. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

