A new module in neural differentiation control: two microRNAs upregulated by retinoic acid, miR-9 and -103, target the differentiation inhibitor ID2
- PMID: 22848373
- PMCID: PMC3405103
- DOI: 10.1371/journal.pone.0040269
A new module in neural differentiation control: two microRNAs upregulated by retinoic acid, miR-9 and -103, target the differentiation inhibitor ID2
Abstract
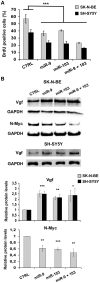
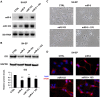
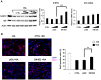
The transcription factor ID2 is an important repressor of neural differentiation strongly implicated in nervous system cancers. MicroRNAs (miRNAs) are increasingly involved in differentiation control and cancer development. Here we show that two miRNAs upregulated on differentiation of neuroblastoma cells--miR-9 and miR-103--restrain ID2 expression by directly targeting the coding sequence and 3' untranslated region of the ID2 encoding messenger RNA, respectively. Notably, the two miRNAs show an inverse correlation with ID2 during neuroblastoma cell differentiation induced by retinoic acid. Overexpression of miR-9 and miR-103 in neuroblastoma cells reduces proliferation and promotes differentiation, as it was shown to occur upon ID2 inhibition. Conversely, an ID2 mutant that cannot be targeted by either miRNA prevents retinoic acid-induced differentiation more efficient than wild-type ID2. These findings reveal a new regulatory module involving two microRNAs upregulated during neural differentiation that directly target expression of the key differentiation inhibitor ID2, suggesting that its alteration may be involved in neural cancer development.
Conflict of interest statement
Figures


References
-
- Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. - PubMed
-
- Ohtani N, Zebedee Z, Huot TJ, Stinson JA, Sugimoto M, et al. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature. 2001;409:1067–1070. - PubMed
-
- Chassot AA, Turchi L, Virolle T, Fitsialos G, Batoz M, et al. Id3 is a novel regulator of p27kip1 mRNA in early G1 phase and is required for cell-cycle progression. Oncogene. 2007;26:5772–5783. - PubMed
-
- Iavarone A, Garg P, Lasorella A, Hsu J, Israel MA. The helix-loop-helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev. 1994;8:1270–1284. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

