Tracking the emergence of a new breed using 49,034 SNP in sheep
- PMID: 22848516
- PMCID: PMC3407242
- DOI: 10.1371/journal.pone.0041508
Tracking the emergence of a new breed using 49,034 SNP in sheep
Abstract
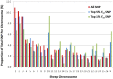
Domestic animals are unique in that they have been organised into managed populations called breeds. The strength of genetic divergence between breeds may vary dependent on the age of the breed, the scenario under which it emerged and the strength of reproductive isolation it has from other breeds. In this study, we investigated the Gulf Coast Native breed of sheep to determine if it contains lines of animals that are sufficiently divergent to be considered separate breeds. Allele sharing and principal component analysis (PCA) using nearly 50,000 SNP loci revealed a clear genetic division that corresponded with membership of either the Florida or Louisiana Native lines. Subsequent analysis aimed to determine if the strength of the divergence exceeded that found between recognised breed pairs. Genotypes from 14 breeds sampled from Europe and Asia were used to obtain estimates of pair-wise population divergence measured as F(ST). The divergence separating the Florida and Louisiana Native (F(ST) = 6.2%) was approximately 50% higher than the average divergence separating breeds developed within the same region of Europe (F(ST) = 4.2%). This strongly indicated that the two Gulf Coast Native lines are sufficiently different to be considered separate breeds. PCA using small SNP sets successfully distinguished between the Florida and Louisiana Native animals, suggesting that allele frequency differences have accumulated across the genome. This is consistent with a population history involving geographic separation and genetic drift. Suggestive evidence was detected for divergence at the poll locus on sheep chromosome 10; however drift at neutral markers has been the largest contributor to the genetic separation observed. These results document the emergence of populations that can be considered separate breeds, with practical consequences for bio-conservation priorities, animal registration and the establishment of separate breed societies.
Conflict of interest statement
Figures



References
-
- Scherf BD (2000) World Watch List for Domestic Animals. Ed. 3. Food and Agriculture Organisation of the United Nations, Rome.
-
- Menotti-Raymond M, David VA, Pflueger SM, Lindblad-Toh K, Wade CM, et al. (2007) Patterns of molecular genetic variation among cat breeds. Genomics 91: 1–11. - PubMed
-
- Lawson Handley LJ, Byrne K, Santucci F, Townsend S, Taylor M, et al. (2007) Genetic structure of European sheep breeds. Heredity 99: 620–631. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

