Investigating the production of foreign membrane proteins in tobacco chloroplasts: expression of an algal plastid terminal oxidase
- PMID: 22848578
- PMCID: PMC3404998
- DOI: 10.1371/journal.pone.0041722
Investigating the production of foreign membrane proteins in tobacco chloroplasts: expression of an algal plastid terminal oxidase
Abstract
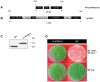
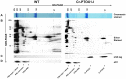
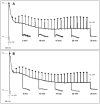
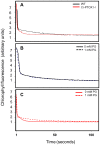
Chloroplast transformation provides an inexpensive, easily scalable production platform for expression of recombinant proteins in plants. However, this technology has been largely limited to the production of soluble proteins. Here we have tested the ability of tobacco chloroplasts to express a membrane protein, namely plastid terminal oxidase 1 from the green alga Chlamydomonas reinhardtii (Cr-PTOX1), which is predicted to function as a plastoquinol oxidase. A homoplastomic plant containing a codon-optimised version of the nuclear gene encoding PTOX1, driven by the 16S rRNA promoter and 5'UTR of gene 10 from phage T7, was generated using a particle delivery system. Accumulation of Cr-PTOX1 was shown by immunoblotting and expression in an enzymatically active form was confirmed by using chlorophyll fluorescence to measure changes in the redox state of the plastoquinone pool in leaves. Growth of Cr-PTOX1 expressing plants was, however, more sensitive to high light than WT. Overall our results confirm the feasibility of using plastid transformation as a means of expressing foreign membrane proteins in the chloroplast.
Conflict of interest statement
Figures



References
-
- Sprenger RR, Jensen ON. Proteomics and the dynamic plasma membrane: Quo Vadis? Proteomics. 2010;10:3997–4011. - PubMed
-
- Bakheet TM, Doig AJ. Properties and identification of human protein drug targets. Bioinformatics. 2009;25:451–457. - PubMed
-
- Lössl AG, Waheed MT. Chloroplast-derived vaccines against human diseases: achievements, challenges and scopes. Plant Biotechnol J. 2011;9:527–539. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

