The phosphorylation-dependent regulation of mitochondrial proteins in stress responses
- PMID: 22848813
- PMCID: PMC3403084
- DOI: 10.1155/2012/931215
The phosphorylation-dependent regulation of mitochondrial proteins in stress responses
Abstract
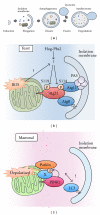
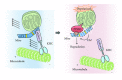
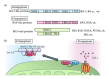
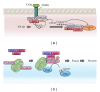
To maintain cellular homeostasis, cells are equipped with precise systems that trigger the appropriate stress responses. Mitochondria not only provide cellular energy but also integrate stress response signaling pathways, including those regulating cell death. Several lines of evidence suggest that the mitochondrial proteins that function in this process, such as Bcl-2 family proteins in apoptosis and phosphoglycerate mutase family member 5 (PGAM5) in necroptosis, are regulated by several kinases. It has also been suggested that the phosphorylation-dependent regulation of mitochondrial fission machinery, dynamin-related protein 1 (Drp1), facilitates appropriate cellular stress responses. However, mitochondria themselves are also damaged by various stresses. To avoid the deleterious effects exerted by damaged mitochondria, cells remove these mitochondria in a selective autophagic degradation process called mitophagy. Interestingly, several kinases, such as PTEN-induced putative kinase 1 (PINK1) in mammals and stress-responsive mitogen-activated protein (MAP) kinases in yeast, have recently been shown to be involved in mitophagy. In this paper, we focus on the phosphorylation-dependent regulation of mitochondrial proteins and discuss the roles of this regulation in the mitochondrial and cellular stress responses.
Figures

References
-
- McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Current Biology. 2006;16(14):R551–R560. - PubMed
-
- Wang X. The expanding role of mitochondria in apoptosis. Genes and Development. 2001;15(22):2922–2933. - PubMed
-
- Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nature Reviews Molecular Cell Biology. 2007;8(11):870–879. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

