Kinetic signatures of microRNA modes of action
- PMID: 22850425
- PMCID: PMC3425779
- DOI: 10.1261/rna.032284.112
Kinetic signatures of microRNA modes of action
Abstract
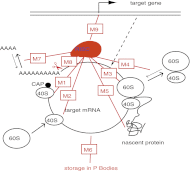
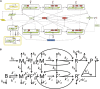
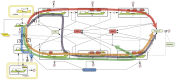
MicroRNAs (miRNAs) are key regulators of all important biological processes, including development, differentiation, and cancer. Although remarkable progress has been made in deciphering the mechanisms used by miRNAs to regulate translation, many contradictory findings have been published that stimulate active debate in this field. Here we contribute to this discussion in three ways. First, based on a comprehensive analysis of the existing literature, we hypothesize a model in which all proposed mechanisms of microRNA action coexist, and where the apparent mechanism that is detected in a given experiment is determined by the relative values of the intrinsic characteristics of the target mRNAs and associated biological processes. Among several coexisting miRNA mechanisms, the one that will effectively be measurable is that which acts on or changes the sensitive parameters of the translation process. Second, we have created a mathematical model that combines nine known mechanisms of miRNA action and estimated the model parameters from the literature. Third, based on the mathematical modeling, we have developed a computational tool for discriminating among different possible individual mechanisms of miRNA action based on translation kinetics data that can be experimentally measured (kinetic signatures). To confirm the discriminatory power of these kinetic signatures and to test our hypothesis, we have performed several computational experiments with the model in which we simulated the coexistence of several miRNA action mechanisms in the context of variable parameter values of the translation.
Figures



References
-
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE 2005. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122: 553–563 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
