The rectal cancer microRNAome--microRNA expression in rectal cancer and matched normal mucosa
- PMID: 22850566
- PMCID: PMC6298229
- DOI: 10.1158/1078-0432.CCR-12-0016
The rectal cancer microRNAome--microRNA expression in rectal cancer and matched normal mucosa
Abstract
Purpose: miRNAs play a prominent role in a variety of physiologic and pathologic biologic processes, including cancer. For rectal cancers, only limited data are available on miRNA expression profiles, whereas the underlying genomic and transcriptomic aberrations have been firmly established. We therefore, aimed to comprehensively map the miRNA expression patterns of this disease.
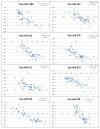
Experimental design: Tumor biopsies and corresponding matched mucosa samples were prospectively collected from 57 patients with locally advanced rectal cancers. Total RNA was extracted, and tumor and mucosa miRNA expression profiles were subsequently established for all patients. The expression of selected miRNAs was validated using semi-quantitative real-time PCR.
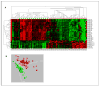
Results: Forty-nine miRNAs were significantly differentially expressed (log(2)-fold difference >0.5 and P < 0.001) between rectal cancer and normal rectal mucosa. The predicted targets for these miRNAs were enriched for the following pathways: Wnt, TGF-beta, mTOR, insulin, mitogen-activated protein kinase, and ErbB signaling. Thirteen of these 49 miRNAs seem to be rectal cancer-specific, and have not been previously reported for colon cancers: miR-492, miR-542-5p, miR-584, miR-483-5p, miR-144, miR-2110, miR-652, miR-375, miR-147b, miR-148a, miR-190, miR-26a/b, and miR-338-3p. Of clinical impact, miR-135b expression correlated significantly with disease-free and cancer-specific survival in an independent multicenter cohort of 116 patients.
Conclusion: This comprehensive analysis of the rectal cancer miRNAome uncovered novel miRNAs and pathways associated with rectal cancer. This information contributes to a detailed view of this disease. Moreover, the identification and validation of miR-135b may help to identify novel molecular targets and pathways for therapeutic exploitation.
©2012 AACR.
Conflict of interest statement
No potential conflicts of interest were disclosed for J.G., M.G., J.C., A.J.S., M.J.D., C.C.H., B.M.G., T.B., and T.R.
R.S., B.K., S.M., and T.L. are former employees of Exiqon A/S.
Figures


References
-
- Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: The impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–36. - PubMed
-
- Sauer R, Becker H, Hohenberger W, Rodel C, Wittekind C, Fietkau R, et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N Engl J Med. 2004;351:1731–40. - PubMed
-
- Sokilde R, Kaczkowski B, Podolska A, Cirera S, Gorodkin J, Moller S, et al. Global microRNA analysis of the NCI-60 cancer cell panel. Mol Cancer Ther. 2011;10:375–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

