Structure and mechanism of purine-binding riboswitches
- PMID: 22850604
- PMCID: PMC3760793
- DOI: 10.1017/S0033583512000078
Structure and mechanism of purine-binding riboswitches
Abstract
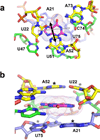
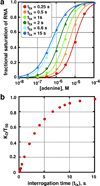
A riboswitch is a non-protein coding sequence capable of directly binding a small molecule effector without the assistance of accessory proteins to regulate expression of the mRNA in which it is embedded. Currently, over 20 different classes of riboswitches have been validated in bacteria with the promise of many more to come, making them an important means of regulating the genome in the bacterial kingdom. Strikingly, half of the known riboswitches recognize effector compounds that contain a purine or related moiety. In the last decade, significant progress has been made to determine how riboswitches specifically recognize these compounds against the background of many other similar cellular metabolites and transduce this signal into a regulatory response. Of the known riboswitches, the purine family containing guanine, adenine and 2'-deoxyguanosine-binding classes are the most extensively studied, serving as a simple and useful paradigm for understanding how these regulatory RNAs function. This review provides a comprehensive summary of the current state of knowledge regarding the structure and mechanism of these riboswitches, as well as insights into how they might be exploited as therapeutic targets and novel biosensors.
Figures
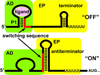
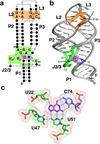
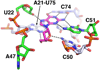
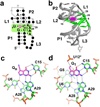
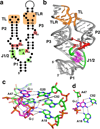
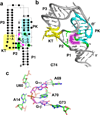
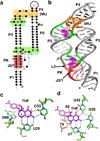
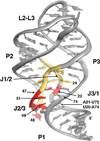


References
-
- Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22(1):3–6. - PubMed
-
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

