Mouse Rif1 is a key regulator of the replication-timing programme in mammalian cells
- PMID: 22850673
- PMCID: PMC3442270
- DOI: 10.1038/emboj.2012.214
Mouse Rif1 is a key regulator of the replication-timing programme in mammalian cells
Abstract
The eukaryotic genome is replicated according to a specific spatio-temporal programme. However, little is known about both its molecular control and biological significance. Here, we identify mouse Rif1 as a key player in the regulation of DNA replication timing. We show that Rif1 deficiency in primary cells results in an unprecedented global alteration of the temporal order of replication. This effect takes place already in the first S-phase after Rif1 deletion and is neither accompanied by alterations in the transcriptional landscape nor by major changes in the biochemical identity of constitutive heterochromatin. In addition, Rif1 deficiency leads to both defective G1/S transition and chromatin re-organization after DNA replication. Together, these data offer a novel insight into the global regulation and biological significance of the replication-timing programme in mammalian cells.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


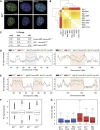

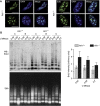



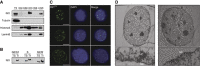
Comment in
-
Rif1 choreographs DNA replication timing.EMBO J. 2012 Sep 12;31(18):3650-2. doi: 10.1038/emboj.2012.238. Epub 2012 Aug 14. EMBO J. 2012. PMID: 22892565 Free PMC article.
References
-
- Anglana M, Apiou F, Bensimon A, Debatisse M (2003) Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell 114: 385–394 - PubMed
-
- Bahtz R, Seidler J, Arnold M, Haselmann-Weiss U, Antony C, Lehmann WD, Hoffmann I (2012) GCP6 is a substrate of Plk4 and required for centriole duplication. J Cell Sci 125(Pt 2): 486–496 - PubMed
-
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J (2005) DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434: 864–870 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

