Rif1 regulates the replication timing domains on the human genome
- PMID: 22850674
- PMCID: PMC3442267
- DOI: 10.1038/emboj.2012.180
Rif1 regulates the replication timing domains on the human genome
Abstract
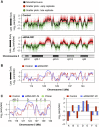
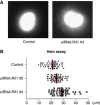
DNA replication is spatially and temporally regulated during S-phase. DNA replication timing is established in early-G1-phase at a point referred to as timing decision point. However, how the genome-wide replication timing domains are established is unknown. Here, we show that Rif1 (Rap1-interacting-factor-1), originally identified as a telomere-binding factor in yeast, is a critical determinant of the replication timing programme in human cells. Depletion of Rif1 results in specific loss of mid-S replication foci profiles, stimulation of initiation events in early-S-phase and changes in long-range replication timing domain structures. Analyses of replication timing show replication of sequences normally replicating early is delayed, whereas that normally replicating late is advanced, suggesting that replication timing regulation is abrogated in the absence of Rif1. Rif1 tightly binds to nuclear-insoluble structures at late-M-to-early-G1 and regulates chromatin-loop sizes. Furthermore, Rif1 colocalizes specifically with the mid-S replication foci. Thus, Rif1 establishes the mid-S replication domains that are restrained from being activated at early-S-phase. Our results indicate that Rif1 plays crucial roles in determining the replication timing domain structures in human cells through regulating higher-order chromatin architecture.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




Comment in
-
Rif1 choreographs DNA replication timing.EMBO J. 2012 Sep 12;31(18):3650-2. doi: 10.1038/emboj.2012.238. Epub 2012 Aug 14. EMBO J. 2012. PMID: 22892565 Free PMC article.
References
-
- Arias EE, Walter JC (2007) Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev 21: 497–518 - PubMed
-
- Buongiorno-Nardelli M, Micheli G, Carri MT, Marilley M (1982) A relationship between replicon size and supercoiled loop domains in the eukaryotic genome. Nature 298: 100–102 - PubMed
-
- Courbet S, Gay S, Arnoult N, Wronka G, Anglana M, Brison O, Debatisse M (2008) Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature 455: 557–560 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

