Structure-based analyses reveal distinct binding sites for Atg2 and phosphoinositides in Atg18
- PMID: 22851171
- PMCID: PMC3442503
- DOI: 10.1074/jbc.M112.397570
Structure-based analyses reveal distinct binding sites for Atg2 and phosphoinositides in Atg18
Abstract
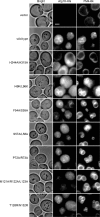
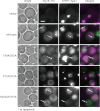
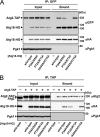
Autophagy is an intracellular degradation system by which cytoplasmic materials are enclosed by an autophagosome and delivered to a lysosome/vacuole. Atg18 plays a critical role in autophagosome formation as a complex with Atg2 and phosphatidylinositol 3-phosphate (PtdIns(3)P). However, little is known about the structure of Atg18 and its recognition mode of Atg2 or PtdIns(3)P. Here, we report the crystal structure of Kluyveromyces marxianus Hsv2, an Atg18 paralog, at 2.6 Å resolution. The structure reveals a seven-bladed β-propeller without circular permutation. Mutational analyses of Atg18 based on the K. marxianus Hsv2 structure suggested that Atg18 has two phosphoinositide-binding sites at blades 5 and 6, whereas the Atg2-binding region is located at blade 2. Point mutations in the loops of blade 2 specifically abrogated autophagy without affecting another Atg18 function, the regulation of vacuolar morphology at the vacuolar membrane. This architecture enables Atg18 to form a complex with Atg2 and PtdIns(3)P in parallel, thereby functioning in the formation of autophagosomes at autophagic membranes.
Figures




References
-
- Mizushima N., Yoshimori T., Ohsumi Y. (2011) The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 - PubMed
-
- Mizushima N. (2007) Autophagy: process and function. Genes Dev. 21, 2861–2873 - PubMed
-
- Mizushima N., Komatsu M. (2011) Autophagy: renovation of cells and tissues. Cell 147, 728–741 - PubMed
-
- Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

