PylSn and the homologous N-terminal domain of pyrrolysyl-tRNA synthetase bind the tRNA that is essential for the genetic encoding of pyrrolysine
- PMID: 22851181
- PMCID: PMC3463324
- DOI: 10.1074/jbc.M112.396754
PylSn and the homologous N-terminal domain of pyrrolysyl-tRNA synthetase bind the tRNA that is essential for the genetic encoding of pyrrolysine
Abstract
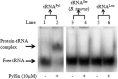
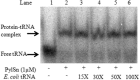
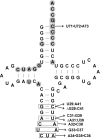
Pyrrolysine is represented by an amber codon in genes encoding proteins such as the methylamine methyltransferases present in some Archaea and Bacteria. Pyrrolysyl-tRNA synthetase (PylRS) attaches pyrrolysine to the amber-suppressing tRNA(Pyl). Archaeal PylRS, encoded by pylS, has a catalytic C-terminal domain but an N-terminal region of unknown function and structure. In Bacteria, homologs of the N- and C-terminal regions of archaeal PylRS are respectively encoded by pylSn and pylSc. We show here that wild type PylS from Methanosarcina barkeri and PylSn from Desulfitobacterium hafniense bind tRNA(Pyl) in EMSA with apparent K(d) values of 0.12 and 0.13 μM, respectively. Truncation of the N-terminal region of PylS eliminated detectable tRNA(Pyl) binding as measured by EMSA, but not catalytic activity. A chimeric protein with PylSn fused to the N terminus of truncated PylS regained EMSA-detectable tRNA(Pyl) binding. PylSn did not bind other D. hafniense tRNAs, nor did the competition by the Escherichia coli tRNA pool interfere with tRNA(Pyl) binding. Further indicating the specificity of PylSn interaction with tRNA(Pyl), substitutions of conserved residues in tRNA(Pyl) in the variable loop, D stem, and T stem and loop had significant impact in binding, whereas those having base changes in the acceptor stem or anticodon stem and loop still retained the ability to complex with PylSn. PylSn and the N terminus of PylS comprise the protein superfamily TIGR03129. The members of this family are not similar to any known RNA-binding protein, but our results suggest their common function involves specific binding of tRNA(Pyl).
Figures





Similar articles
-
Recognition of pyrrolysine tRNA by the Desulfitobacterium hafniense pyrrolysyl-tRNA synthetase.Nucleic Acids Res. 2007;35(4):1270-8. doi: 10.1093/nar/gkl1151. Epub 2007 Jan 31. Nucleic Acids Res. 2007. PMID: 17267409 Free PMC article.
-
Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality.Nature. 2009 Feb 26;457(7233):1163-7. doi: 10.1038/nature07611. Epub 2008 Dec 31. Nature. 2009. PMID: 19118381 Free PMC article.
-
The amino-terminal domain of pyrrolysyl-tRNA synthetase is dispensable in vitro but required for in vivo activity.FEBS Lett. 2007 Jul 10;581(17):3197-203. doi: 10.1016/j.febslet.2007.06.004. Epub 2007 Jun 12. FEBS Lett. 2007. PMID: 17582401 Free PMC article.
-
tRNAPyl: Structure, function, and applications.RNA Biol. 2018;15(4-5):441-452. doi: 10.1080/15476286.2017.1356561. Epub 2017 Sep 13. RNA Biol. 2018. PMID: 28837402 Free PMC article. Review.
-
The direct genetic encoding of pyrrolysine.Curr Opin Microbiol. 2005 Dec;8(6):706-12. doi: 10.1016/j.mib.2005.10.009. Epub 2005 Oct 26. Curr Opin Microbiol. 2005. PMID: 16256420 Review.
Cited by
-
Enhancing the performance of a mutant pyrrolysyl-tRNA synthetase to create a highly versatile eukaryotic cell-free protein synthesis tool.Sci Rep. 2023 Sep 14;13(1):15236. doi: 10.1038/s41598-023-42198-8. Sci Rep. 2023. PMID: 37709815 Free PMC article.
-
Two-Tier Screening Platform for Directed Evolution of Aminoacyl-tRNA Synthetases with Enhanced Stop Codon Suppression Efficiency.Chembiochem. 2017 Jun 19;18(12):1109-1116. doi: 10.1002/cbic.201700039. Epub 2017 May 16. Chembiochem. 2017. PMID: 28383180 Free PMC article.
-
Engineering aminoacyl-tRNA synthetases for use in synthetic biology.Enzymes. 2020;48:351-395. doi: 10.1016/bs.enz.2020.06.004. Epub 2020 Sep 8. Enzymes. 2020. PMID: 33837709 Free PMC article.
-
Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.ACS Chem Biol. 2022 Dec 16;17(12):3470-3477. doi: 10.1021/acschembio.2c00640. Epub 2022 Nov 17. ACS Chem Biol. 2022. PMID: 36395426 Free PMC article.
-
Crystal structures reveal an elusive functional domain of pyrrolysyl-tRNA synthetase.Nat Chem Biol. 2017 Dec;13(12):1261-1266. doi: 10.1038/nchembio.2497. Epub 2017 Oct 16. Nat Chem Biol. 2017. PMID: 29035363 Free PMC article.
References
-
- Atkins J. F., Gesteland R. (2002) The 22nd amino acid. Science 296, 1409–1410 - PubMed
-
- Krzycki J. A. (2004) Function of genetically encoded pyrrolysine in corrinoid-dependent methylamine methyltransferases. Curr. Opin. Chem. Biol. 8, 484–491 - PubMed
-
- Hao B., Gong W., Ferguson T. K., James C. M., Krzycki J. A., Chan M. K. (2002) A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science 296, 1462–1466 - PubMed
-
- Soares J. A., Zhang L., Pitsch R. L., Kleinholz N. M., Jones R. B., Wolff J. J., Amster J., Green-Church K. B., Krzycki J. A. (2005) The residue mass of l-pyrrolysine in three distinct methylamine methyltransferases. J. Biol. Chem. 280, 36962–36969 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

