Mediator promotes CENP-a incorporation at fission yeast centromeres
- PMID: 22851695
- PMCID: PMC3457525
- DOI: 10.1128/MCB.00374-12
Mediator promotes CENP-a incorporation at fission yeast centromeres
Erratum in
- Mol Cell Biol. 2012 Dec;32(24):5151
-
Correction for Carlsten et al., "Mediator Promotes CENP-A Incorporation at Fission Yeast Centromeres".Mol Cell Biol. 2021 Jun 23;41(7):e0017921. doi: 10.1128/MCB.00179-21. Epub 2021 Jun 23. Mol Cell Biol. 2021. PMID: 34160264 Free PMC article. No abstract available.
Abstract
At Schizosaccharomyces pombe centromeres, heterochromatin formation is required for de novo incorporation of the histone H3 variant CENP-A(Cnp1), which in turn directs kinetochore assembly and ultimately chromosome segregation during mitosis. Noncoding RNAs (ncRNAs) transcribed by RNA polymerase II (Pol II) directs heterochromatin formation through not only the RNA interference (RNAi) machinery but also RNAi-independent RNA processing factors. Control of centromeric ncRNA transcription is therefore a key factor for proper centromere function. We here demonstrate that Mediator directs ncRNA transcription and regulates centromeric heterochromatin formation in fission yeast. Mediator colocalizes with Pol II at centromeres, and loss of the Mediator subunit Med20 causes a dramatic increase in pericentromeric transcription and desilencing of the core centromere. As a consequence, heterochromatin formation is impaired via both the RNAi-dependent and -independent pathways, resulting in loss of CENP-A(Cnp1) from the core centromere, a defect in kinetochore function, and a severe chromosome segregation defect. Interestingly, the increased centromeric transcription observed in med20Δ cells appears to directly block CENP-A(Cnp1) incorporation since inhibition of Pol II transcription can suppress the observed phenotypes. Our data thus identify Mediator as a crucial regulator of ncRNA transcription at fission yeast centromeres and add another crucial layer of regulation to centromere function.
Figures

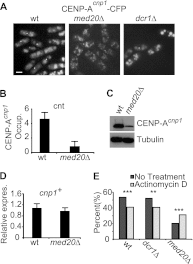

References
-
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9: 218–233 - PubMed
-
- Appelgren H, Kniola B, Ekwall K. 2003. Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells. J. Cell Sci. 116: 4035–4042 - PubMed
-
- Bannister AJ, et al. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 - PubMed
-
- Basi G, Schmid E, Maundrell K. 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123: 131–136 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
