Modulation of toxin production by the flagellar regulon in Clostridium difficile
- PMID: 22851750
- PMCID: PMC3457548
- DOI: 10.1128/IAI.00224-12
Modulation of toxin production by the flagellar regulon in Clostridium difficile
Abstract
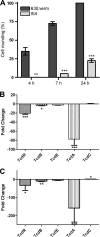
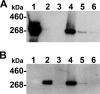
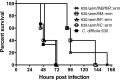
We show in this study that toxin production in Clostridium difficile is altered in cells which can no longer form flagellar filaments. The impact of inactivation of fliC, CD0240, fliF, fliG, fliM, and flhB-fliR flagellar genes upon toxin levels in culture supernatants was assessed using cell-based cytotoxicity assay, proteomics, immunoassay, and immunoblotting approaches. Each of these showed that toxin levels in supernatants were significantly increased in a fliC mutant compared to that in the C. difficile 630 parent strain. In contrast, the toxin levels in supernatants secreted from other flagellar mutants were significantly reduced compared with that in the parental C. difficile 630 strain. Transcriptional analysis of the pathogenicity locus genes (tcdR, tcdB, tcdE, and tcdA) revealed a significant increase of all four genes in the fliC mutant strain, while transcription of all four genes was significantly reduced in fliM, fliF, fliG, and flhB-fliR mutants. These results demonstrate that toxin transcription in C. difficile is modulated by the flagellar regulon. More significantly, mutant strains showed a corresponding change in virulence compared to the 630 parent strain when tested in a hamster model of C. difficile infection. This is the first demonstration of differential flagellum-related transcriptional regulation of toxin production in C. difficile and provides evidence for elaborate regulatory networks for virulence genes in C. difficile.
Figures




References
-
- Akerlund T, Svenungsson BB, Lagergren A, Burman LG. 2006. Correlation of disease severity with fecal toxin levels in patients with Clostridium difficile-associated diarrhea and distribution of PCR ribotypes and toxin yields in vitro of corresponding isolates. J. Clin. Microbiol. 44:353–358 - PMC - PubMed
-
- Ananthakrishnan AN. 2011. Clostridium difficile infection: epidemiology, risk factors and management. Nat. Rev. Gastroenterol. Hepatol. 8:17–26 - PubMed
-
- Antunes A, Martin-Verstraete I, Dupuy B. 2011. CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol. Microbiol. 79:882–899 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

