SNPs as Supplements in Simple Kinship Analysis or as Core Markers in Distant Pairwise Relationship Tests: When Do SNPs Add Value or Replace Well-Established and Powerful STR Tests?
- PMID: 22851936
- PMCID: PMC3375139
- DOI: 10.1159/000338857
SNPs as Supplements in Simple Kinship Analysis or as Core Markers in Distant Pairwise Relationship Tests: When Do SNPs Add Value or Replace Well-Established and Powerful STR Tests?
Abstract
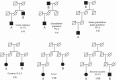
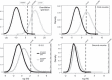
BACKGROUND: Genetic tests for kinship testing routinely reach likelihoods that provide virtual proof of the claimed relationship by typing microsatellites-commonly consisting of 12-15 standard forensic short tandem repeats (STRs). Single nucleotide polymorphisms (SNPs) have also been applied to kinship testing but these binary markers are required in greater numbers than multiple-allele STRs. However SNPs offer certain advantageous characteristics not found in STRs, including, much higher mutational stability, good performance typing highly degraded DNA, and the ability to be readily up-scaled to very high marker numbers reaching over a million loci. This article outlines kinship testing applications where SNPs markedly improve the genetic data obtained. In particular we explore the minimum number of SNPs that will be required to confirm pairwise relationship claims in deficient pedigrees that typify missing persons' identification or war grave investigations where commonly few surviving relatives are available for comparison and the DNA is highly degraded. METHODS: We describe the application of SNPs alongside STRs when incomplete profiles or allelic instability in STRs create ambiguous results, we review the use of high density SNP arrays when the relationship claim is very distant, and we outline simulations of kinship analyses with STRs supplemented with SNPs in order to estimate the practical limit of pairwise relationships that can be differentiated from random unrelated pairs from the same population. RESULTS: The minimum number of SNPs for robust statistical inference of parent-offspring relationships through to those of second cousins (S-3-3) is estimated for both simple, single multiplex SNP sets and for subsets of million-SNP arrays. CONCLUSIONS: There is considerable scope for resolving ambiguous STR results and for improving the statistical power of kinship analysis by adding small-scale SNP sets but where the pedigree is deficient the pairwise relationships must be relatively close. For more distant relationships it is possible to reduce chip-based SNP arrays from the million+ markers down to ∼7,000. However, such numbers indicate that current genotyping approaches will not be able to deliver sufficient data to resolve distant pairwise relationships from the limited DNA typical of the most challenging identification cases.
Hintergrund: Genetische Tests für Abstammungsgutachten erreichen normalerweise das zum Nachweis einer Verwandtschaft erforderliche Wahrscheinlichkeitsniveau durch die Typisierung von in der Forensik etablierten Mikrosatelliten, welche häufig aus 12–15 kurzen, hintereinander auftretenden Sequenzwiederholungen (STRs, engl. short tandem repeats) bestehen. Einzel-nukleotid-Polymorphismen (SNPs, engl. single nucleotide polymorphism) werden ebenfalls in der Verwandtschaftsanalyse eingesetzt, wobei diese binären Marker aber in einer größeren Anzahl als STRs mit multiplen Allelen erforderlich sind. Jedoch bieten SNPs einige vorteilhafte Eigenschaften die STRs nicht aufweisen: größere Mutationsstabilität, gute Analysierbarkeit bei der Typisierung von stark degradierter DNA und die Fähigkeit zu unkomplizierten Erweiterung des Marker-Sets bis zu einer Anzahl von über einer Million. Dieser Artikel beschreibt Anwendungen von Abstammungsgutachten, in denen SNPs deutlich die erhaltenen genetischen Daten verbessern. Insbesondere untersuchen wir die minimale Anzahl von erforderlichen SNPs zur Bestätigung paarweiser Verwandtschaft in Defizienzfällen, die oftmals bei der Identifizierung vermisster Personen oder der Untersuchung von Kriegsgräbern auftreten, bei denen meist nur wenige Angehörige zu Vergleichszwecken zur Verfügung stehen und zudem die DNA stark degradiert ist.
Methoden: Wir beschreiben die simultane Anwendung von SNPs und STRs, wenn inkomplette Profile oder allelische Instabilität der STRs zu unklaren Ergebnissen führen, erläutern den Gebrauch von hochauflösenden SNP-Ana-lysesystemen, wenn das zu untersuchende Verwandtschaftsverhältnis weit auseinander liegt, und schildern die Simulation von paarweisen Verwandtschaftsuntersuchungen unter Anwendung von STRs und SNPs zur Abschätzung der Limitation bei der Differenzierung zwischen paarweisen Verwandtschaftsverhältnissen von zufälligen, unverwandten Paaren aus derselben Population.
Ergebnisse: Die minimale Anzahl von SNPs für eine gesicherte Rekonstruktion von Eltern-Kind-Beziehungen bis hin zu solchen für Vettern zweiten Grades wird für einfache Multiplex-SNP-Sets und für Zusammenstellungen von Millionen von SNPs abgeschätzt.
Schlussfolgerung: Es gibt eine große Bandbreite an Möglichkeiten, um durch die Hinzunahme einer begrenzten Anzahl von SNPs unklare STR-Ergebnisse zu lösen oder die statistische Aussagekraft von Abstammungsbegutachtungen zu verbessern, wobei jedoch die paarweise Beziehung relativ eng sein muss. Für weiter entfernte paarweise Verwandtschaftsverhältnisse können Chip-basierte SNP-Analyse-systeme von über 1 Millionen Marker auf ∼7,000 reduziert werden. Jedoch zeigen diese Zahlen, dass die momentan zur Verfügung stehenden Genotypi-sierungssysteme aufgrund der normalerweise in komplizierten Identifikationsfällen limitierten DNA nicht in der Lage sind, genügend Daten zu liefern, um entfernte paarweise Verwandtschaftsverhältnisse aufzuklären.
Figures



Similar articles
-
A 472-SNP panel for pairwise kinship testing of second-degree relatives.Forensic Sci Int Genet. 2018 May;34:178-185. doi: 10.1016/j.fsigen.2018.02.019. Epub 2018 Mar 2. Forensic Sci Int Genet. 2018. PMID: 29510334
-
Improved pairwise kinship analysis using massively parallel sequencing.Forensic Sci Int Genet. 2019 Jan;38:77-85. doi: 10.1016/j.fsigen.2018.10.006. Epub 2018 Oct 10. Forensic Sci Int Genet. 2019. PMID: 30368110
-
Complex kinship analysis with a combination of STRs, SNPs, and indels.Forensic Sci Int Genet. 2022 Nov;61:102749. doi: 10.1016/j.fsigen.2022.102749. Epub 2022 Jul 20. Forensic Sci Int Genet. 2022. PMID: 35939875
-
"New turns from old STaRs": enhancing the capabilities of forensic short tandem repeat analysis.Electrophoresis. 2014 Nov;35(21-22):3173-87. doi: 10.1002/elps.201400095. Epub 2014 Jul 16. Electrophoresis. 2014. PMID: 24888494 Review.
-
SNPs and STRs in forensic medicine. A strategy for kinship evaluation.Arch Med Sadowej Kryminol. 2017;67(3):226-240. doi: 10.5114/amsik.2017.73194. Arch Med Sadowej Kryminol. 2017. PMID: 29460612 Review. English.
Cited by
-
Improved second-degree kinship analysis using the FGID forensic four-in-one DNA typing kit.Int J Legal Med. 2025 Sep;139(5):2159-2163. doi: 10.1007/s00414-025-03491-5. Epub 2025 Apr 10. Int J Legal Med. 2025. PMID: 40208275
-
SNP typing using the HID-Ion AmpliSeq™ Identity Panel in a southern Chinese population.Int J Legal Med. 2018 Jul;132(4):997-1006. doi: 10.1007/s00414-017-1706-3. Epub 2017 Oct 18. Int J Legal Med. 2018. PMID: 29046953
-
Potential forensic use of a 33 X-InDel panel in the Argentinean population.Int J Legal Med. 2017 Jan;131(1):107-112. doi: 10.1007/s00414-016-1399-z. Epub 2016 Jun 9. Int J Legal Med. 2017. PMID: 27282766 Review.
-
100 Years after von Dungern & Hirschfeld: Kinship Investigation from Blood Groups to SNPs.Transfus Med Hemother. 2012 Jun;39(3):161-162. doi: 10.1159/000339263. Epub 2012 May 15. Transfus Med Hemother. 2012. PMID: 22851930 Free PMC article. No abstract available.
-
A novel 193-plex MPS panel integrating STRs and SNPs highlights the application value of forensic genetics in individual identification and paternity testing.Hum Genet. 2024 Mar;143(3):371-383. doi: 10.1007/s00439-024-02658-1. Epub 2024 Mar 18. Hum Genet. 2024. PMID: 38499885
References
-
- Phillips C, Fondevila M, García-Magarinos M, Rodriguez A, Salas A, Carrecedo A, Lareu MV. Resolving relationship tests that show ambiguous STR results using autosomal SNPs as supplementary markers. Forensic Sci Int Genet. 2008;2:198–204. - PubMed
-
- Pereira R, Phillips C, Alves C, Amorim A, Carracedo A, Gusmão L. A new multiplex for human identification using insertion/deletion polymorphisms. Electrophoresis. 2009;30:3682–3690. - PubMed
-
- Børsting C, Morling N. Mutations and/or relatives? Six case work examples where 49 autosomal SNPs were used as supplementary markers. Forensic Sci Int Genet. 2011;5:236–241. - PubMed
-
- Børsting C, Morling N. Re-investigations of six unusual paternity cases by typing of autosomal single nucleotide polymorphisms. Transfusion. 2012;52:425–430. - PubMed
-
- Lareu MV, García-Magariños M, Phillips C, Quintela I, Carracedo Á, Salas A. Analysis of a claimed distant relationship in a deficient pedigree using high density SNP data. Forensic Sci Int Genet. 2012;6:350–353. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

