Complete nucleotide sequence of watermelon chlorotic stunt virus originating from Oman
- PMID: 22852046
- PMCID: PMC3407900
- DOI: 10.3390/v4071169
Complete nucleotide sequence of watermelon chlorotic stunt virus originating from Oman
Abstract
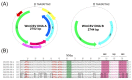
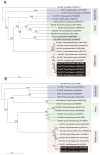
Watermelon chlorotic stunt virus (WmCSV) is a bipartite begomovirus (genus Begomovirus, family Geminiviridae) that causes economic losses to cucurbits, particularly watermelon, across the Middle East and North Africa. Recently squash (Cucurbita moschata) grown in an experimental field in Oman was found to display symptoms such as leaf curling, yellowing and stunting, typical of a begomovirus infection. Sequence analysis of the virus isolated from squash showed 97.6-99.9% nucleotide sequence identity to previously described WmCSV isolates for the DNA A component and 93-98% identity for the DNA B component. Agrobacterium-mediated inoculation to Nicotiana benthamiana resulted in the development of symptoms fifteen days post inoculation. This is the first bipartite begomovirus identified in Oman. Overall the Oman isolate showed the highest levels of sequence identity to a WmCSV isolate originating from Iran, which was confirmed by phylogenetic analysis. This suggests that WmCSV present in Oman has been introduced from Iran. The significance of this finding is discussed.
Keywords: Begomovirus; Oman; WmCSV; squash.
Figures


Similar articles
-
Molecular characterization of watermelon chlorotic stunt virus (WmCSV) from Palestine.Viruses. 2014 Jun 20;6(6):2444-62. doi: 10.3390/v6062444. Viruses. 2014. PMID: 24956181 Free PMC article.
-
Watermelon chlorotic stunt virus (WmCSV): a serious disease threatening watermelon production in Jordan.Virus Genes. 2011 Aug;43(1):79-89. doi: 10.1007/s11262-011-0594-8. Epub 2011 Mar 12. Virus Genes. 2011. PMID: 21399920
-
Analysis of watermelon chlorotic stunt virus and tomato leaf curl Palampur virus mixed and pseudo-recombination infections.Virus Genes. 2015 Dec;51(3):408-16. doi: 10.1007/s11262-015-1250-5. Epub 2015 Oct 3. Virus Genes. 2015. PMID: 26433951
-
Disease complex associated with begomoviruses infecting squash and cucumber in Saudi Arabia.Cell Mol Biol (Noisy-le-grand). 2024 Nov 27;70(11):58-63. doi: 10.14715/cmb/2024.70.11.8. Cell Mol Biol (Noisy-le-grand). 2024. PMID: 39707780
-
Characterization of a synergistic interaction between two cucurbit-infecting begomoviruses: Squash leaf curl virus and Watermelon chlorotic stunt virus.Phytopathology. 2011 Feb;101(2):281-9. doi: 10.1094/PHYTO-06-10-0159. Phytopathology. 2011. PMID: 21219130
Cited by
-
Molecular and biological characterization of Chilli leaf curl virus and associated Tomato leaf curl betasatellite infecting tobacco in Oman.Virol J. 2019 Nov 9;16(1):131. doi: 10.1186/s12985-019-1235-4. Virol J. 2019. PMID: 31706358 Free PMC article.
-
Identification of the Begomoviruses Squash Leaf Curl Virus and Watermelon Chlorotic Stunt Virus in Various Plant Samples in North America.Viruses. 2021 Apr 30;13(5):810. doi: 10.3390/v13050810. Viruses. 2021. PMID: 33946382 Free PMC article.
-
Infectivity of cloned begomoviral DNAs: an appraisal.Virusdisease. 2019 Mar;30(1):13-21. doi: 10.1007/s13337-018-0453-5. Epub 2018 May 3. Virusdisease. 2019. PMID: 31143828 Free PMC article.
-
Molecular characterization of watermelon chlorotic stunt virus (WmCSV) from Palestine.Viruses. 2014 Jun 20;6(6):2444-62. doi: 10.3390/v6062444. Viruses. 2014. PMID: 24956181 Free PMC article.
-
Insights into the Incidence of Watermelon chlorotic stunt virus Causing Yellowing Disease of Watermelon in Western and Southwestern Regions of Saudi Arabia.Plant Pathol J. 2018 Oct;34(5):426-434. doi: 10.5423/PPJ.OA.04.2018.0059. Epub 2018 Oct 1. Plant Pathol J. 2018. PMID: 30369852 Free PMC article.
References
-
- Brown J.K., Fauquet C.M., Briddon R.W., Zerbini M., Moriones E., Navas-Castillo J. Geminiviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy - Ninth Report of the International Committee on Taxonomy of Viruses. Associated Press, Elsevier Inc.; London, UK: 2012. pp. 351–373.
-
- Moffat A.S. Geminiviruses emerge as serious crop threat. Science. 1999;286:1835. doi: 10.1126/science.286.5446.1835. - DOI
-
- Mansoor S., Briddon R.W., Bull S.E., Bedford I.D., Bashir A., Hussain M., Saeed M., Zafar M.Y., Malik K.A., Fauquet C., et al. Cotton leaf curl disease is associated with multiple monopartite begomoviruses supported by single DNA β. Arch. Virol. 2003;148:1969–1986. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

