Comparison of gene expression profiling in sarcomas and mesenchymal stem cells identifies tumorigenic pathways in chemically induced rat sarcoma model
- PMID: 22852096
- PMCID: PMC3407640
- DOI: 10.5402/2012/909453
Comparison of gene expression profiling in sarcomas and mesenchymal stem cells identifies tumorigenic pathways in chemically induced rat sarcoma model
Abstract
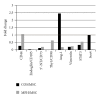
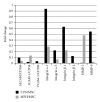
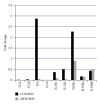
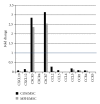
Mesenchymal stem cells (MSCs) are believed to be the cell of origin for most sarcomas including osteosarcoma and malignant fibrous histiocytoma (MFH/UPS). To identify the signaling pathways involved in sarcoma pathogenesis, we compared gene expression profiles in rat osteosarcoma and MFH cells with those in syngeneic rat MSCs. Analysis of genes that characterize MSCs such as CD44, CD105, CD73, and CD90 showed higher expression in MSCs compared to sarcomas. Pathways involved in focal and cell adhesion, cytokine-cytokine receptors, extracellular matrix receptors, chemokines, and Wnt signaling were down-regulated in both sarcomas. Meanwhile, DNA replication, cell cycle, mismatch repair, Hedgehog signaling, and metabolic pathways were upregulated in both sarcomas. Downregulation of p21(Cip1) and higher expression of CDK4-cyclinD1 and CDK2-cyclinE could accelerate cell cycle in sarcomas. The current study indicated that these rat sarcomas could be a good model for their human counterparts and will provide the further insights into the molecular pathways and mechanisms involved in sarcoma pathogenesis.
Figures

Similar articles
-
Rat malignant fibrous histiocytoma (MFH)-derived cloned cell lines (MT-8 and MT-9) show different differentiation in mesenchymal stem cell lineage.Exp Toxicol Pathol. 2015 Oct;67(10):499-507. doi: 10.1016/j.etp.2015.07.004. Epub 2015 Jul 21. Exp Toxicol Pathol. 2015. PMID: 26208870
-
Mesenchymal stem cells promote tumor engraftment and metastatic colonization in rat osteosarcoma model.Int J Oncol. 2012 Jan;40(1):163-9. doi: 10.3892/ijo.2011.1220. Epub 2011 Oct 3. Int J Oncol. 2012. PMID: 21971610
-
Mesenchymal stem cell transformation and sarcoma genesis.Clin Sarcoma Res. 2013 Jul 23;3(1):10. doi: 10.1186/2045-3329-3-10. Clin Sarcoma Res. 2013. PMID: 23880362 Free PMC article.
-
Global protein-expression profiling for reclassification of malignant fibrous histiocytoma.Biochim Biophys Acta. 2015 Jun;1854(6):696-701. doi: 10.1016/j.bbapap.2014.08.012. Epub 2014 Aug 28. Biochim Biophys Acta. 2015. PMID: 25173742 Review.
-
Sarcoma Stem Cell Heterogeneity.Adv Exp Med Biol. 2019;1123:95-118. doi: 10.1007/978-3-030-11096-3_7. Adv Exp Med Biol. 2019. PMID: 31016597 Review.
Cited by
-
Lost miRNA surveillance of Notch, IGFR pathway--road to sarcomagenesis.Tumour Biol. 2014 Jan;35(1):483-92. doi: 10.1007/s13277-013-1068-5. Tumour Biol. 2014. PMID: 23959473
-
β-Catenin transcriptional activity is minimal in canine osteosarcoma and its targeted inhibition results in minimal changes to cell line behaviour.Vet Comp Oncol. 2016 Jun;14(2):e4-e16. doi: 10.1111/vco.12077. Epub 2013 Nov 21. Vet Comp Oncol. 2016. PMID: 24256430 Free PMC article.
-
Histologic and genetic advances in refining the diagnosis of "undifferentiated pleomorphic sarcoma".Cancers (Basel). 2013 Feb 22;5(1):218-33. doi: 10.3390/cancers5010218. Cancers (Basel). 2013. PMID: 24216705 Free PMC article.
-
CDK/CCN and CDKI alterations for cancer prognosis and therapeutic predictivity.Biomed Res Int. 2014;2014:361020. doi: 10.1155/2014/361020. Epub 2014 Jan 29. Biomed Res Int. 2014. PMID: 24605326 Free PMC article. Review.
-
Tumourigenic canine osteosarcoma cell lines associated with frizzled-6 up-regulation and enhanced side population cell frequency.Vet Comp Oncol. 2017 Mar;15(1):78-93. doi: 10.1111/vco.12138. Epub 2015 Feb 16. Vet Comp Oncol. 2017. PMID: 25689105 Free PMC article.
References
-
- Fletcher CDM, Unni KK, Mertens F, et al. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Soft Tissue and Bone. Lyon, France: IARC Press; 2002. Conventional osteosarcoma.
-
- Raymond AK, Ayala AG, Knuutila S. Conventional osteosarcoma. In: Fletcher CDM, Unni KK, Mertens F, editors. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Soft Tissue and Bone. Lyon, France: IARC Press; 2002. pp. 264–270.
-
- Fletcher CDM. The evolving classification of soft tissue tumours: an update based on the new WHO classification. Histopathology. 2006;48(1):3–12. - PubMed
-
- Nakayama R, Nemoto T, Takahashi H, et al. Gene expression analysis of soft tissue sarcomas: characterization and reclassification of malignant fibrous histiocytoma. Modern Pathology. 2007;20(7):749–759. - PubMed
-
- Judson I. State-of-the-art approach in selective curable tumours: soft tissue sarcoma. Annals of Oncology. 2008;19(7):vii166–vii169. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

