Widespread existence of cytosine methylation in yeast DNA measured by gas chromatography/mass spectrometry
- PMID: 22852529
- PMCID: PMC4706227
- DOI: 10.1021/ac301727c
Widespread existence of cytosine methylation in yeast DNA measured by gas chromatography/mass spectrometry
Abstract
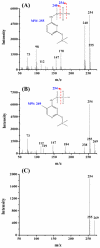
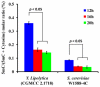
DNA methylation is one of the major epigenetic modifications and has been involved in a number of biological processes in mammalian cells. Yeast is widely used as a model organism for studying cell metabolism, cell cycle regulation, and signal transduction. However, it remains controversial whether methylated cytosine (5-methylcytosine, 5mC) exists in the yeast genome. In the current study, we developed a highly sensitive method based on gas chromatography/mass spectrometry (GC/MS) and systematically examined the incidence of 5mC in 19 yeast strains, which represent 16 yeast species. Our results showed that DNA methylation is widespread in yeast and the genome-wide DNA methylation of the studied yeast strains ranged from 0.014 to 0.364%, which were 1 to 2 orders of magnitude lower than that in mammalian cells (i.e., 3-8%). Furthermore, we found that the 5mC content in yeast varied considerably at different growth stages and DNA methylation inhibitor 5-azacytidine could induce a decrease in genome-wide DNA methylation as that in mammalian cells. The demonstration of the universal presence of DNA cytosine methylation in yeast constituted the first and essential step toward understanding the functions of this methylation in yeast.
Figures


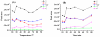
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

