Replica exchanging self-guided Langevin dynamics for efficient and accurate conformational sampling
- PMID: 22852596
- PMCID: PMC3416874
- DOI: 10.1063/1.4737094
Replica exchanging self-guided Langevin dynamics for efficient and accurate conformational sampling
Abstract
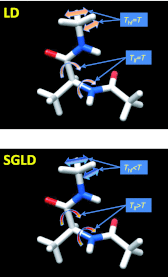
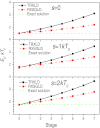
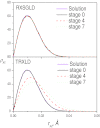
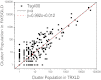
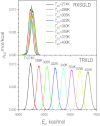
This work presents a replica exchanging self-guided Langevin dynamics (RXSGLD) simulation method for efficient conformational searching and sampling. Unlike temperature-based replica exchanging simulations, which use high temperatures to accelerate conformational motion, this method uses self-guided Langevin dynamics (SGLD) to enhance conformational searching without the need to elevate temperatures. A RXSGLD simulation includes a series of SGLD simulations, with simulation conditions differing in the guiding effect and/or temperature. These simulation conditions are called stages and the base stage is one with no guiding effect. Replicas of a simulation system are simulated at the stages and are exchanged according to the replica exchanging probability derived from the SGLD partition function. Because SGLD causes less perturbation on conformational distribution than high temperatures, exchanges between SGLD stages have much higher probabilities than those between different temperatures. Therefore, RXSGLD simulations have higher conformational searching ability than temperature based replica exchange simulations. Through three example systems, we demonstrate that RXSGLD can generate target canonical ensemble distribution at the base stage and achieve accelerated conformational searching. Especially for large systems, RXSGLD has remarkable advantages in terms of replica exchange efficiency, conformational searching ability, and system size extensiveness.
Figures








References
-
- Wu X. and Brooks B. R., Chem. Phys. Lett. 381, 512 (2003).10.1016/j.cplett.2003.10.013 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

