Direct simulation of early-stage Sec-facilitated protein translocation
- PMID: 22852862
- PMCID: PMC3431202
- DOI: 10.1021/ja3034526
Direct simulation of early-stage Sec-facilitated protein translocation
Abstract
Direct simulations reveal key mechanistic features of early-stage protein translocation and membrane integration via the Sec-translocon channel. We present a novel computational protocol that combines non-equilibrium growth of the nascent protein with microsecond timescale molecular dynamics trajectories. Analysis of multiple, long timescale simulations elucidates molecular features of protein insertion into the translocon, including signal-peptide docking at the translocon lateral gate (LG), large lengthscale conformational rearrangement of the translocon LG helices, and partial membrane integration of hydrophobic nascent-protein sequences. Furthermore, the simulations demonstrate the role of specific molecular interactions in the regulation of protein secretion, membrane integration, and integral membrane protein topology. Salt-bridge contacts between the nascent-protein N-terminus, cytosolic translocon residues, and phospholipid head groups are shown to favor conformations of the nascent protein upon early-stage insertion that are consistent with the Type II (N(cyt)/C(exo)) integral membrane protein topology, and extended hydrophobic contacts between the nascent protein and the membrane lipid bilayer are shown to stabilize configurations that are consistent with the Type III (N(exo)/C(cyt)) topology. These results provide a detailed, mechanistic basis for understanding experimentally observed correlations between integral membrane protein topology, translocon mutagenesis, and nascent-protein sequence.
Figures

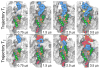

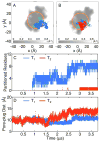
 , that partition into the membrane during the insertion simulations. (D) The time-evolution of the pore-plug distance in the insertion simulations.
, that partition into the membrane during the insertion simulations. (D) The time-evolution of the pore-plug distance in the insertion simulations.

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

