Profiling the resting venom gland of the scorpion Tityus stigmurus through a transcriptomic survey
- PMID: 22853446
- PMCID: PMC3444934
- DOI: 10.1186/1471-2164-13-362
Profiling the resting venom gland of the scorpion Tityus stigmurus through a transcriptomic survey
Abstract
Background: The scorpion Tityus stigmurus is widely distributed in Northeastern Brazil and known to cause severe human envenoming, inducing pain, hyposthesia, edema, erythema, paresthesia, headaches and vomiting. The present study uses a transcriptomic approach to characterize the gene expression profile from the non-stimulated venom gland of Tityus stigmurus scorpion.
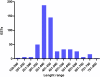
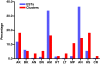
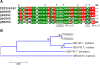
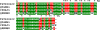
Results: A cDNA library was constructed and 540 clones were sequenced and grouped into 153 clusters, with one or more ESTs (expressed sequence tags). Forty-one percent of ESTs belong to recognized toxin-coding sequences, with transcripts encoding antimicrobial toxins (AMP-like) being the most abundant, followed by alfa KTx- like, beta KTx-like, beta NaTx-like and alfa NaTx-like. Our analysis indicated that 34% of the transcripts encode "other possible venom molecules", which correspond to anionic peptides, hypothetical secreted peptides, metalloproteinases, cystein-rich peptides and lectins. Fifteen percent of ESTs are similar to cellular transcripts. Sequences without good matches corresponded to 11%.
Conclusions: This investigation provides the first global view of gene expression of the venom gland from Tityus stigmurus under resting conditions. This approach enables characterization of a large number of venom gland component molecules, which belong either to known or non yet described types of venom peptides and proteins from the Buthidae family.
Figures







References
-
- Polis GA. The biology of scorpions. Stanford University Press, California; 1990.
-
- Saúde FN, editor. Book Manual de Diagnóstico e Tratamento de Acidentes por Animais Peçonhentos. FUNASA, City; 2001.
-
- Lira-da-Silva RM, Amorim AM, Brazil TK. Poisonous sting by Tityus stigmurus (Scorpiones; Buthidae) in the state of Bahia, Brazil. Rev Soc Bras Med Trop. 2000;33:239–245. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

