Trapping DNA near a solid-state nanopore
- PMID: 22853913
- PMCID: PMC3400781
- DOI: 10.1016/j.bpj.2012.06.008
Trapping DNA near a solid-state nanopore
Abstract
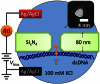
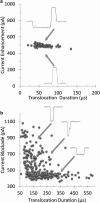
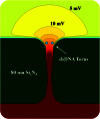
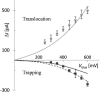
We demonstrate that voltage-biased solid-state nanopores can transiently localize DNA in an electrolyte solution. A double-stranded DNA (dsDNA) molecule is trapped when the electric field near the nanopore attracts and immobilizes a non-end segment of the molecule across the nanopore orifice without inducing a folded molecule translocation. In this demonstration of the phenomenon, the ionic current through the nanopore decreases when the dsDNA molecule is trapped by the nanopore. By contrast, a translocating dsDNA molecule under the same conditions causes an ionic current increase. We also present finite-element modeling results that predict this behavior for the conditions of the experiment.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
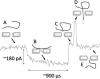
Similar articles
-
DNA motion induced by electrokinetic flow near an Au coated nanopore surface as voltage controlled gate.Nanotechnology. 2015 Feb 13;26(6):065502. doi: 10.1088/0957-4484/26/6/065502. Epub 2015 Jan 22. Nanotechnology. 2015. PMID: 25611963 Free PMC article.
-
Ionic Current Fluctuation and Orientation of Tetrahedral DNA Nanostructures in a Solid-State Nanopore.Small. 2022 Mar;18(12):e2107237. doi: 10.1002/smll.202107237. Epub 2022 Jan 29. Small. 2022. PMID: 35092143
-
Surface Charge Density-Dependent DNA Capture through Polymer Planar Nanopores.ACS Appl Mater Interfaces. 2018 Nov 28;10(47):40927-40937. doi: 10.1021/acsami.8b14423. Epub 2018 Nov 13. ACS Appl Mater Interfaces. 2018. PMID: 30371050
-
Multi-resolution simulation of DNA transport through large synthetic nanostructures.Phys Chem Chem Phys. 2022 Feb 2;24(5):2706-2716. doi: 10.1039/d1cp04589j. Phys Chem Chem Phys. 2022. PMID: 35050282 Free PMC article. Review.
-
Slowing and controlling the translocation of DNA in a solid-state nanopore.Nanoscale. 2012 Feb 21;4(4):1068-77. doi: 10.1039/c1nr11201e. Epub 2011 Nov 14. Nanoscale. 2012. PMID: 22081018 Free PMC article. Review.
Cited by
-
Solid-state nanopore hydrodynamics and transport.Biomicrofluidics. 2019 Jan 30;13(1):011301. doi: 10.1063/1.5083913. eCollection 2019 Jan. Biomicrofluidics. 2019. PMID: 30867871 Free PMC article.
-
Voltage-driven translocation behaviors of IgG molecule through nanopore arrays.Nanoscale Res Lett. 2013 May 15;8(1):229. doi: 10.1186/1556-276X-8-229. Nanoscale Res Lett. 2013. PMID: 23676116 Free PMC article.
-
Tracking single-particle dynamics via combined optical and electrical sensing.Sci Rep. 2013;3:1855. doi: 10.1038/srep01855. Sci Rep. 2013. PMID: 23685401 Free PMC article.
-
Enhanced Discriminability of Viral Vectors in Viscous Nanopores.Small Methods. 2025 Jul;9(7):e2401321. doi: 10.1002/smtd.202401321. Epub 2025 Jan 2. Small Methods. 2025. PMID: 39743980 Free PMC article.
-
Molecule-hugging graphene nanopores.Proc Natl Acad Sci U S A. 2013 Jul 23;110(30):12192-6. doi: 10.1073/pnas.1220012110. Epub 2013 Jul 8. Proc Natl Acad Sci U S A. 2013. PMID: 23836648 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

