PIDD death-domain phosphorylation by ATM controls prodeath versus prosurvival PIDDosome signaling
- PMID: 22854598
- PMCID: PMC3444620
- DOI: 10.1016/j.molcel.2012.06.024
PIDD death-domain phosphorylation by ATM controls prodeath versus prosurvival PIDDosome signaling
Abstract
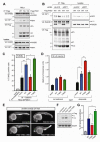
Biochemical evidence implicates the death-domain (DD) protein PIDD as a molecular switch capable of signaling cell survival or death in response to genotoxic stress. PIDD activity is determined by binding-partner selection at its DD: whereas recruitment of RIP1 triggers prosurvival NF-κB signaling, recruitment of RAIDD activates proapoptotic caspase-2 via PIDDosome formation. However, it remains unclear how interactor selection, and thus fate decision, is regulated at the PIDD platform. We show that the PIDDosome functions in the "Chk1-suppressed" apoptotic response to DNA damage, a conserved ATM/ATR-caspase-2 pathway antagonized by Chk1. In this pathway, ATM phosphorylates PIDD on Thr788 within the DD. This phosphorylation is necessary and sufficient for RAIDD binding and caspase-2 activation. Conversely, nonphosphorylatable PIDD fails to bind RAIDD or activate caspase-2, and engages prosurvival RIP1 instead. Thus, ATM phosphorylation of the PIDD DD enables a binary switch through which cells elect to survive or die upon DNA injury.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures





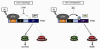
Comment in
-
Janus-faced PIDD: a sensor for DNA damage-induced cell death or survival?Mol Cell. 2012 Sep 14;47(5):667-8. doi: 10.1016/j.molcel.2012.08.019. Mol Cell. 2012. PMID: 22980457
References
-
- Cuenin S, Tinel A, Janssens S, Tschopp J. p53-induced protein with a death domain (PIDD) isoforms differentially activate nuclear factor-kappaB and caspase-2 in response to genotoxic stress. Oncogene. 2008;27:387–396. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

