The antisense RNA As1_flv4 in the Cyanobacterium Synechocystis sp. PCC 6803 prevents premature expression of the flv4-2 operon upon shift in inorganic carbon supply
- PMID: 22854963
- PMCID: PMC3460422
- DOI: 10.1074/jbc.M112.391755
The antisense RNA As1_flv4 in the Cyanobacterium Synechocystis sp. PCC 6803 prevents premature expression of the flv4-2 operon upon shift in inorganic carbon supply
Abstract
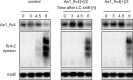
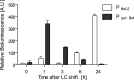
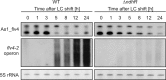
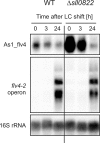
The functional relevance of natural cis-antisense transcripts is mostly unknown. Here we have characterized the association of three antisense RNAs and one intergenically encoded noncoding RNA with an operon that plays a crucial role in photoprotection of photosystem II under low carbon conditions in the cyanobacterium Synechocystis sp. PCC 6803. Cyanobacteria show strong gene expression dynamics in response to a shift of cells from high carbon to low levels of inorganic carbon (C(i)), but the regulatory mechanisms are poorly understood. Among the most up-regulated genes in Synechocystis are flv4, sll0218, and flv2, which are organized in the flv4-2 operon. The flavodiiron proteins encoded by this operon open up an alternative electron transfer route, likely starting from the Q(B) site in photosystem II, under photooxidative stress conditions. Our expression analysis of cells shifted from high carbon to low carbon demonstrated an inversely correlated transcript accumulation of the flv4-2 operon mRNA and one antisense RNA to flv4, designated as As1_flv4. Overexpression of As1_flv4 led to a decrease in flv4-2 mRNA. The promoter activity of as1_flv4 was transiently stimulated by C(i) limitation and negatively regulated by the AbrB-like transcription regulator Sll0822, whereas the flv4-2 operon was positively regulated by the transcription factor NdhR. The results indicate that the tightly regulated antisense RNA As1_flv4 establishes a transient threshold for flv4-2 expression in the early phase after a change in C(i) conditions. Thus, it prevents unfavorable synthesis of the proteins from the flv4-2 operon.
Figures


Similar articles
-
Flavodiiron protein Flv2/Flv4-related photoprotective mechanism dissipates excitation pressure of PSII in cooperation with phycobilisomes in Cyanobacteria.Plant Physiol. 2014 Feb;164(2):805-18. doi: 10.1104/pp.113.231969. Epub 2013 Dec 23. Plant Physiol. 2014. PMID: 24367022 Free PMC article.
-
Flavodiiron proteins in oxygenic photosynthetic organisms: photoprotection of photosystem II by Flv2 and Flv4 in Synechocystis sp. PCC 6803.PLoS One. 2009;4(4):e5331. doi: 10.1371/journal.pone.0005331. Epub 2009 Apr 24. PLoS One. 2009. PMID: 19390625 Free PMC article.
-
Operon flv4-flv2 provides cyanobacterial photosystem II with flexibility of electron transfer.Plant Cell. 2012 May;24(5):1952-71. doi: 10.1105/tpc.111.094417. Epub 2012 May 8. Plant Cell. 2012. PMID: 22570444 Free PMC article.
-
Riboregulators and the role of Hfq in photosynthetic bacteria.RNA Biol. 2014;11(5):413-26. doi: 10.4161/rna.28035. Epub 2014 Feb 10. RNA Biol. 2014. PMID: 24651049 Free PMC article. Review.
-
Cyanobacterial Oxygenic Photosynthesis is Protected by Flavodiiron Proteins.Life (Basel). 2015 Mar 9;5(1):716-43. doi: 10.3390/life5010716. Life (Basel). 2015. PMID: 25761262 Free PMC article. Review.
Cited by
-
Genetic and genomic analysis of RNases in model cyanobacteria.Photosynth Res. 2015 Oct;126(1):171-83. doi: 10.1007/s11120-015-0076-2. Epub 2015 Jan 17. Photosynth Res. 2015. PMID: 25595545 Free PMC article.
-
Iron deprivation in Synechocystis: inference of pathways, non-coding RNAs, and regulatory elements from comprehensive expression profiling.G3 (Bethesda). 2012 Dec;2(12):1475-95. doi: 10.1534/g3.112.003863. Epub 2012 Dec 1. G3 (Bethesda). 2012. PMID: 23275872 Free PMC article.
-
Proteomic approaches in research of cyanobacterial photosynthesis.Photosynth Res. 2015 Oct;126(1):47-70. doi: 10.1007/s11120-014-0050-4. Epub 2014 Oct 31. Photosynth Res. 2015. PMID: 25359503 Review.
-
Lighting the way: Compelling open questions in photosynthesis research.Plant Cell. 2024 Oct 3;36(10):3914-3943. doi: 10.1093/plcell/koae203. Plant Cell. 2024. PMID: 39038210 Free PMC article.
-
The primary transcriptome of the fast-growing cyanobacterium Synechococcus elongatus UTEX 2973.Biotechnol Biofuels. 2018 Aug 4;11:218. doi: 10.1186/s13068-018-1215-8. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 30127850 Free PMC article.
References
-
- De Lucia F., Dean C. (2011) Long non-coding RNAs and chromatin regulation. Curr. Opin. Plant Biol. 14, 168–173 - PubMed
-
- Zhang X., Xia J., Lii Y. E., Barrera-Figueroa B. E., Zhou X., Gao S., Lu L., Niu D., Chen Z., Leung C., Wong T., Zhang H., Guo J., Li Y., Liu R., Liang W., Zhu J. K., Zhang W., Jin H. (2012) Genome-wide analysis of plant nat-siRNAs reveals insights into their distribution, biogenesis and function. Genome Biol. 13, R20. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

