DERP6 (ELP5) and C3ORF75 (ELP6) regulate tumorigenicity and migration of melanoma cells as subunits of Elongator
- PMID: 22854966
- PMCID: PMC3463322
- DOI: 10.1074/jbc.M112.402727
DERP6 (ELP5) and C3ORF75 (ELP6) regulate tumorigenicity and migration of melanoma cells as subunits of Elongator
Abstract
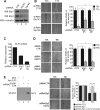
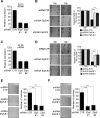
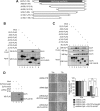
The Elongator complex is composed of 6 subunits (Elp1-Elp6) and promotes RNAPII transcript elongation through histone acetylation in the nucleus as well as tRNA modification in the cytoplasm. This acetyltransferase complex directly or indirectly regulates numerous biological processes ranging from exocytosis and resistance to heat shock in yeast to cell migration and neuronal differentiation in higher eukaryotes. The identity of human ELP1 through ELP4 has been reported but human ELP5 and ELP6 have remained uncharacterized. Here, we report that DERP6 (ELP5) and C3ORF75 (ELP6) encode these subunits of human Elongator. We further investigated the importance and function of these two subunits by a combination of biochemical analysis and cellular assays. Our results show that DERP6/ELP5 is required for the integrity of Elongator and directly connects ELP3 to ELP4. Importantly, the migration and tumorigenicity of melanoma-derived cells are significantly decreased upon Elongator depletion through ELP1 or ELP3. Strikingly, DERP6/ELP5 and C3ORF75/ELP6-depleted melanoma cells have similar defects, further supporting the idea that DERP6/ELP5 and C3ORF75/ELP6 are essential for Elongator function. Together, our data identify DERP6/ELP5 and C3ORF75/ELP6 as key players for migration, invasion and tumorigenicity of melanoma cells, as integral subunits of Elongator.
Figures




References
-
- Otero G., Fellows J., Li Y., de Bizemont T., Dirac A. M., Gustafsson C. M., Erdjument-Bromage H., Tempst P., Svejstrup J. Q. (1999) Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3, 109–118 - PubMed
-
- Hawkes N. A., Otero G., Winkler G. S., Marshall N., Dahmus M. E., Krappmann D., Scheidereit C., Thomas C. L., Schiavo G., Erdjument-Bromage H., Tempst P., Svejstrup J. Q. (2002) Purification and characterization of the human elongator complex. J. Biol. Chem. 277, 3047–3052 - PubMed
-
- Wittschieben B. O., Otero G., de Bizemont T., Fellows J., Erdjument-Bromage H., Ohba R., Li Y., Allis C. D., Tempst P., Svejstrup J. Q. (1999) A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 4, 123–128 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

