Interaction between isolated transcriptional activation domains of Sp1 revealed by heteronuclear magnetic resonance
- PMID: 22855260
- PMCID: PMC3526990
- DOI: 10.1002/pro.2137
Interaction between isolated transcriptional activation domains of Sp1 revealed by heteronuclear magnetic resonance
Abstract
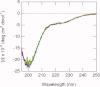
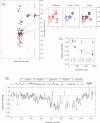
The promoter-specific transcription factor Sp1 is expressed ubiquitously, and plays a primary role in the regulation of the expression of many genes. Domains A and B located in the N-terminal half of the protein are characterized by glutamine-rich (Q-rich) sequences. These Q-rich domains have been shown to be involved in the interaction between Sp1 and different classes of nuclear proteins, such as TATA-binding protein associated factors. Furthermore, the self-association of Sp1 via Q-rich domains is also important for the regulation of transcriptional activity. It has been considered that an Sp1 molecule bound to a "distal" GC-box synergistically interacts with another Sp1 molecule at a "proximal" binding site. Although the formation of multimers via Q-rich domains seems functionally important for Sp1, little is known about the structural and physicochemical nature of the interaction between Q-rich domains. We analyzed the structural details of isolated glutamine-rich B (QB) domains of Sp1 by circular dichroism (CD), analytical ultracentrifugation, and heteronuclear magnetic resonance spectroscopy (NMR). We found the isolated QB domains to be disordered under all conditions examined. Nevertheless, a detailed analysis of NMR spectra clearly indicated interaction between the domains. In particular, the C-terminal half was responsible for the self-association. Furthermore, analytical ultracentrifugation demonstrated weak but significant interaction between isolated QB domains. The self-association between QB domains would be responsible, at least in part, for the formation of multimers by full-length Sp1 molecules that has been proposed to occur during transcriptional activation.
Copyright © 2012 The Protein Society.
Figures



Similar articles
-
Interaction between intrinsically disordered regions in transcription factors Sp1 and TAF4.Protein Sci. 2016 Nov;25(11):2006-2017. doi: 10.1002/pro.3013. Epub 2016 Aug 24. Protein Sci. 2016. PMID: 27515574 Free PMC article.
-
Identification of heteromolecular binding sites in transcription factors Sp1 and TAF4 using high-resolution nuclear magnetic resonance spectroscopy.Protein Sci. 2017 Nov;26(11):2280-2290. doi: 10.1002/pro.3287. Epub 2017 Sep 27. Protein Sci. 2017. PMID: 28857320 Free PMC article.
-
A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation.Proc Natl Acad Sci U S A. 1994 Jan 4;91(1):192-6. doi: 10.1073/pnas.91.1.192. Proc Natl Acad Sci U S A. 1994. PMID: 8278363 Free PMC article.
-
Transcriptional regulation by zinc-finger proteins Sp1 and MAZ involves interactions with the same cis-elements.Int J Mol Med. 2003 May;11(5):547-53. Int J Mol Med. 2003. PMID: 12684688 Review.
-
SP1 transcription factors in male germ cell development and differentiation.Mol Cell Endocrinol. 2007 May 30;270(1-2):1-7. doi: 10.1016/j.mce.2007.03.001. Epub 2007 Mar 12. Mol Cell Endocrinol. 2007. PMID: 17462816 Review.
Cited by
-
Interaction between intrinsically disordered regions in transcription factors Sp1 and TAF4.Protein Sci. 2016 Nov;25(11):2006-2017. doi: 10.1002/pro.3013. Epub 2016 Aug 24. Protein Sci. 2016. PMID: 27515574 Free PMC article.
-
A novel mode of interaction between intrinsically disordered proteins.Biophys Physicobiol. 2020 Jul 3;17:86-93. doi: 10.2142/biophysico.BSJ-2020012. eCollection 2020. Biophys Physicobiol. 2020. PMID: 33194509 Free PMC article. Review.
-
Interaction networks of prion, prionogenic and prion-like proteins in budding yeast, and their role in gene regulation.PLoS One. 2014 Jun 27;9(6):e100615. doi: 10.1371/journal.pone.0100615. eCollection 2014. PLoS One. 2014. PMID: 24972093 Free PMC article.
-
Letter to the Editor: A still unresolved mystery in the interaction between intrinsically disordered proteins: How do they recognize multiple target proteins? A commentary on "No folding upon binding of intrinsically disordered proteins: Still interesting but not unique and novel. by Sigalov, A. B., Biophysics and Physicobiology 17, 156-158 (2020). DOI: 10.2142/biophysico.BSJ-2020025".Biophys Physicobiol. 2020 Dec 18;17:159-160. doi: 10.2142/biophysico.BSJ-2020028. eCollection 2020. Biophys Physicobiol. 2020. PMID: 33447499 Free PMC article. No abstract available.
-
Bex1 is essential for ciliogenesis and harbours biomolecular condensate-forming capacity.BMC Biol. 2022 Feb 10;20(1):42. doi: 10.1186/s12915-022-01246-x. BMC Biol. 2022. PMID: 35144600 Free PMC article.
References
-
- Kavurma MM, Santiago FS, Bonfoco E, Khachigian LM. Sp1 phosphorylation regulates apoptosis via extracellular FasL-Fas engagement. J Biol Chem. 2001;275:4964–4971. - PubMed
-
- Courey AJ, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. - PubMed
-
- Kadonaga JT, Carner KR, Masiarz FR, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079–1090. - PubMed
-
- Elrod-Erickson M, Pabo CO. Binding studies with mutants of Zif268. J Biol Chem. 1999;274:19281–19285. - PubMed
-
- Wilkins RC, Lis JT. DNA distortion and multimerization: novel functions of the glutamine-rich domain of GAGA factor. J Mol Biol. 1999;285:515–525. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

