The mechanism of the converter domain rotation in the recovery stroke of myosin motor protein
- PMID: 22855405
- PMCID: PMC3486948
- DOI: 10.1002/prot.24155
The mechanism of the converter domain rotation in the recovery stroke of myosin motor protein
Abstract
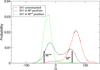
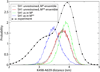
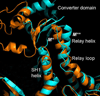
Upon ATP binding, myosin motor protein is found in two alternative conformations, prerecovery state M* and postrecovery state M**. The transition from one state to the other, known as the recovery stroke, plays a key role in the myosin functional cycle. Despite much recent research, the microscopic details of this transition remain elusive. A critical step in the recovery stroke is the rotation of the converter domain from "up" position in prerecovery state to "down" position in postrecovery state that leads to the swing of the lever arm attached to it. In this work, we demonstrate that the two rotational states of the converter domain are determined by the interactions within a small structural motif in the force-generating region of the protein that can be accurately modeled on computers using atomic representation and explicit solvent. Our simulations show that the transition between the two states is controlled by a small helix (SH1) located next to the relay helix and relay loop. A small translation in the position of SH1 away from the relay helix is seen to trigger the transition from "up" state to "down" state. The transition is driven by a cluster of hydrophobic residues I687, F487, and F506 that make significant contributions to the stability of both states. The proposed mechanism agrees well with the available structural and mutational studies.
Copyright © 2012 Wiley Periodicals, Inc.
Figures








References
-
- Lymn RW, Taylor EW. Mechanism of Adenosine Triphosphate Hydrolysis by Actomyosin. Biochemistry. 1971;10(25):4617–4624. - PubMed
-
- Malnasi-Csizmadia A, Pearson DS, Kovacs M, Woolley RJ, Geeves MA, Bagshaw CR. Kinetic resolution of a conformational transition and the ATP hydrolysis step using relaxation methods with a Dictyostelium myosin II mutant containing a single tryptophan residue. Biochemistry. 2001;40(42):12727–12737. - PubMed
-
- Sweeney HL, Houdusse A. Structural and Functional Insights into the Myosin Motor Mechanism. Annual Review of Biophysics, Vol 39. 2010;39:539–557. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

