Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA
- PMID: 22855488
- PMCID: PMC3457165
- DOI: 10.1128/JVI.01700-12
Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA
Abstract
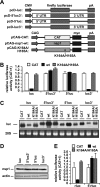
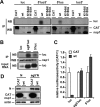
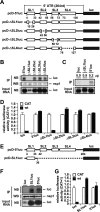
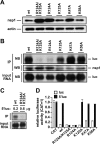
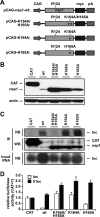
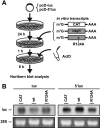
Severe acute respiratory syndrome (SARS) coronavirus (SCoV) is an enveloped virus containing a single-stranded, positive-sense RNA genome. Nine mRNAs carrying a set of common 5' and 3' untranslated regions (UTR) are synthesized from the incoming viral genomic RNA in cells infected with SCoV. A nonstructural SCoV nsp1 protein causes a severe translational shutoff by binding to the 40S ribosomal subunits. The nsp1-40S ribosome complex further induces an endonucleolytic cleavage near the 5'UTR of host mRNA. However, the mechanism by which SCoV viral proteins are efficiently produced in infected cells in which host protein synthesis is impaired by nsp1 is unknown. In this study, we investigated the role of the viral UTRs in evasion of the nsp1-mediated shutoff. Luciferase activities were significantly suppressed in cells expressing nsp1 together with the mRNA carrying a luciferase gene, while nsp1 failed to suppress luciferase activities of the mRNA flanked by the 5'UTR of SCoV. An RNA-protein binding assay and RNA decay assay revealed that nsp1 bound to stem-loop 1 (SL1) in the 5'UTR of SCoV RNA and that the specific interaction with nsp1 stabilized the mRNA carrying SL1. Furthermore, experiments using an SCoV replicon system showed that the specific interaction enhanced the SCoV replication. The specific interaction of nsp1 with SL1 is an important strategy to facilitate efficient viral gene expression in infected cells, in which nsp1 suppresses host gene expression. Our data indicate a novel mechanism of viral gene expression control by nsp1 and give new insight into understanding the pathogenesis of SARS.
Figures
Similar articles
-
SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage.PLoS Pathog. 2011 Dec;7(12):e1002433. doi: 10.1371/journal.ppat.1002433. Epub 2011 Dec 8. PLoS Pathog. 2011. PMID: 22174690 Free PMC article.
-
Characterization of nsp1 Binding to the Viral RNA Leader Sequence of Severe Acute Respiratory Syndrome Coronavirus.Biochemistry. 2024 May 21;63(10):1235-1240. doi: 10.1021/acs.biochem.4c00078. Epub 2024 May 8. Biochemistry. 2024. PMID: 38718213 Free PMC article.
-
Putative cis-acting stem-loops in the 5' untranslated region of the severe acute respiratory syndrome coronavirus can substitute for their mouse hepatitis virus counterparts.J Virol. 2006 Nov;80(21):10600-14. doi: 10.1128/JVI.00455-06. Epub 2006 Aug 18. J Virol. 2006. PMID: 16920822 Free PMC article.
-
Mechanisms of Coronavirus Nsp1-Mediated Control of Host and Viral Gene Expression.Cells. 2021 Feb 2;10(2):300. doi: 10.3390/cells10020300. Cells. 2021. PMID: 33540583 Free PMC article. Review.
-
The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing.Adv Virus Res. 2016;96:59-126. doi: 10.1016/bs.aivir.2016.08.008. Epub 2016 Sep 14. Adv Virus Res. 2016. PMID: 27712628 Free PMC article. Review.
Cited by
-
Viral Respiratory Pathogens and Lung Injury.Clin Microbiol Rev. 2021 Mar 31;34(3):e00103-20. doi: 10.1128/CMR.00103-20. Print 2021 Jun 16. Clin Microbiol Rev. 2021. PMID: 33789928 Free PMC article. Review.
-
SARS and MERS: recent insights into emerging coronaviruses.Nat Rev Microbiol. 2016 Aug;14(8):523-34. doi: 10.1038/nrmicro.2016.81. Epub 2016 Jun 27. Nat Rev Microbiol. 2016. PMID: 27344959 Free PMC article. Review.
-
All Domains of SARS-CoV-2 nsp1 Determine Translational Shutoff and Cytotoxicity of the Protein.J Virol. 2023 Mar 30;97(3):e0186522. doi: 10.1128/jvi.01865-22. Epub 2023 Feb 27. J Virol. 2023. PMID: 36847528 Free PMC article.
-
Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1.Virology. 2016 Feb;489:252-68. doi: 10.1016/j.virol.2015.12.010. Epub 2016 Jan 14. Virology. 2016. PMID: 26773386 Free PMC article.
-
Two-amino acids change in the nsp4 of SARS coronavirus abolishes viral replication.Virology. 2017 Oct;510:165-174. doi: 10.1016/j.virol.2017.07.019. Epub 2017 Jul 21. Virology. 2017. PMID: 28738245 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

