Genome-wide analysis reveals recurrent structural abnormalities of TP63 and other p53-related genes in peripheral T-cell lymphomas
- PMID: 22855598
- PMCID: PMC5070713
- DOI: 10.1182/blood-2012-03-419937
Genome-wide analysis reveals recurrent structural abnormalities of TP63 and other p53-related genes in peripheral T-cell lymphomas
Abstract
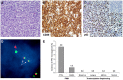
Peripheral T-cell lymphomas (PTCLs) are aggressive malignancies of mature T lymphocytes with 5-year overall survival rates of only ∼ 35%. Improvement in outcomes has been stymied by poor understanding of the genetics and molecular pathogenesis of PTCL, with a resulting paucity of molecular targets for therapy. We developed bioinformatic tools to identify chromosomal rearrangements using genome-wide, next-generation sequencing analysis of mate-pair DNA libraries and applied these tools to 16 PTCL patient tissue samples and 6 PTCL cell lines. Thirteen recurrent abnormalities were identified, of which 5 involved p53-related genes (TP53, TP63, CDKN2A, WWOX, and ANKRD11). Among these abnormalities were novel TP63 rearrangements encoding fusion proteins homologous to ΔNp63, a dominant-negative p63 isoform that inhibits the p53 pathway. TP63 rearrangements were seen in 11 (5.8%) of 190 PTCLs and were associated with inferior overall survival; they also were detected in 2 (1.2%) of 164 diffuse large B-cell lymphomas. As TP53 mutations are rare in PTCL compared with other malignancies, our findings suggest that a constellation of alternate genetic abnormalities may contribute to disruption of p53-associated tumor suppressor function in PTCL.
Figures




References
-
- Savage KJ, Chhanabhai M, Gascoyne RD, Connors JM. Characterization of peripheral T-cell lymphomas in a single North American institution by the WHO classification. Ann Oncol. 2004;15(10):1467–1475. - PubMed
-
- Savage KJ, Harris NL, Vose JM, et al. ALK- anaplastic large-cell lymphoma is clinically and immunophenotypically different from both ALK+ ALCL and peripheral T-cell lymphoma, not otherwise specified: report from the International Peripheral T-Cell Lymphoma Project. Blood. 2008;111(12):5496–5504. - PubMed
-
- WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. In: Swerdlow S, Campo E, Harris N, et al., editors; Bosman F, Jaffe E, Lakhani S, Ohgaki H, editors. World Health Organization Classification of Tumours. 4 Ed. Lyon, France: International Agency for Research on Cancer; 2008.
-
- Foss FM, Zinzani PL, Vose JM, Gascoyne RD, Rosen ST, Tobinai K. Peripheral T-cell lymphoma. Blood. 2011;117(25):6756–6767. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

