Host co-factors of the retrovirus-like transposon Ty1
- PMID: 22856544
- PMCID: PMC3522557
- DOI: 10.1186/1759-8753-3-12
Host co-factors of the retrovirus-like transposon Ty1
Abstract
Background: Long-terminal repeat (LTR) retrotransposons have complex modes of mobility involving reverse transcription of their RNA genomes in cytoplasmic virus-like particles (VLPs) and integration of the cDNA copies into the host genome. The limited coding capacity of retrotransposons necessitates an extensive reliance on host co-factors; however, it has been challenging to identify co-factors that are required for endogenous retrotransposon mobility because retrotransposition is such a rare event.
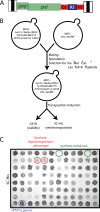
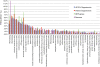
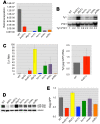
Results: To circumvent the low frequency of Ty1 LTR-retrotransposon mobility in Saccharomyces cerevisiae, we used iterative synthetic genetic array (SGA) analysis to isolate host mutations that reduce retrotransposition. Query strains that harbor a chromosomal Ty1his3AI reporter element and either the rtt101Δ or med1Δ mutation, both of which confer a hypertransposition phenotype, were mated to 4,847 haploid ORF deletion strains. Retrotransposition was measured in the double mutant progeny, and a set of 275 ORF deletions that suppress the hypertransposition phenotypes of both rtt101Δ and med1Δ were identified. The corresponding set of 275 retrotransposition host factors (RHFs) includes 45 previously identified Ty1 or Ty3 co-factors. More than half of the RHF genes have statistically robust human homologs (E < 1 x 10-10). The level of unintegrated Ty1 cDNA in 181 rhfΔ single mutants was altered <2-fold, suggesting that the corresponding co-factors stimulate retrotransposition at a step after cDNA synthesis. However, deletion of 43 RHF genes, including specific ribosomal protein and ribosome biogenesis genes and RNA degradation, modification and transport genes resulted in low Ty1 cDNA levels. The level of Ty1 Gag but not RNA was reduced in ribosome biogenesis mutants bud21Δ, hcr1Δ, loc1Δ, and puf6Δ.
Conclusion: Ty1 retrotransposition is dependent on multiple co-factors acting at different steps in the replication cycle. Human orthologs of these RHFs are potential, or in a few cases, presumptive HIV-1 co-factors in human cells. RHF genes whose absence results in decreased Ty1 cDNA include characterized RNA metabolism and modification genes, consistent with their having roles in early steps in retrotransposition such as expression, nuclear export, translation, localization, or packaging of Ty1 RNA. Our results suggest that Bud21, Hcr1, Loc1, and Puf6 promote efficient synthesis or stability of Ty1 Gag.
Figures

References
-
- Curcio MJ, Derbyshire KM. The outs and ins of transposition: from mu to kangaroo. Nat Rev Mol Cell Biol. 2003;4:865–877. - PubMed
-
- Boeke J, Sandmeyer S. In: Yeast transposable elements. Broach J, Jones E, Pringle J, editor. Cold Spring Harbor Laboratory Press, NY; 1991. The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae; pp. 193–261.
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

